Expression, purification, and characterization of transmembrane protein homogentisate solanesyltransferase
- PMID: 38451307
- PMCID: PMC10920428
- DOI: 10.1007/s00253-024-13094-6
Expression, purification, and characterization of transmembrane protein homogentisate solanesyltransferase
Abstract
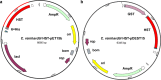
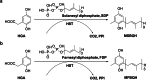
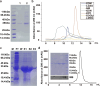
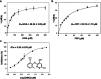
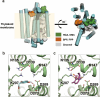
Homogentisate solanesyltransferase (HST) is a crucial enzyme in the plastoquinone biosynthetic pathway and has recently emerged as a promising target for herbicides. In this study, we successfully expressed and purified a stable and highly pure form of seven times transmembrane protein Chlamydomonas reinhardtii HST (CrHST). The final yield of CrHST protein obtained was 12.2 mg per liter of M9 medium. We evaluated the inhibitory effect on CrHST using Des-Morpholinocarbony Cyclopyrimorate (DMC) and found its IC50 value to be 3.63 ± 0.53 μM, indicating significant inhibitory potential. Additionally, we investigated the substrate affinity of CrHST with two substrates, determining the Km values as 22.76 ± 1.70 μM for FPP and 48.54 ± 3.89 μM for HGA. Through sequence alignment analyses and three-dimensional structure predictions, we identified conserved amino acid residues forming the active cavity in the enzyme. The results from molecular docking and binding energy calculations indicate that DMC has a greater binding affinity with HST compared to HGA. These findings represent substantial progress in understanding CrHST's properties and potential for herbicide development. KEY POINTS: • First high-yield transmembrane CrHST protein via E. coli system • Preliminarily identified active cavity composition via activity testing • Determined substrate and inhibitor modes via molecular docking.
Keywords: Bacterial expression system; Enzyme activity; Homogentisate solanesyltransferase; Membrane protein; Molecular docking.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
In vivo and in vitro evidence for the inhibition of homogentisate solanesyltransferase by cyclopyrimorate.Pest Manag Sci. 2020 Oct;76(10):3389-3394. doi: 10.1002/ps.5698. Epub 2019 Dec 24. Pest Manag Sci. 2020. PMID: 31773889
-
Action mechanism of bleaching herbicide cyclopyrimorate, a novel homogentisate solanesyltransferase inhibitor.J Pestic Sci. 2018 Nov 20;43(4):233-239. doi: 10.1584/jpestics.D18-008. J Pestic Sci. 2018. PMID: 30479543 Free PMC article.
-
Characterization of homogentisate prenyltransferases involved in plastoquinone-9 and tocochromanol biosynthesis.FEBS Lett. 2006 Oct 2;580(22):5357-62. doi: 10.1016/j.febslet.2006.09.002. Epub 2006 Sep 12. FEBS Lett. 2006. PMID: 16989822
-
Catalytic reactions of the homogentisate prenyl transferase involved in plastoquinone-9 biosynthesis.J Biol Chem. 2010 Jun 11;285(24):18191-8. doi: 10.1074/jbc.M110.117929. Epub 2010 Apr 16. J Biol Chem. 2010. PMID: 20400515 Free PMC article.
-
The Chlamydomonas reinhardtii gtr gene encoding the tetrapyrrole biosynthetic enzyme glutamyl-trna reductase: structure of the gene and properties of the expressed enzyme.Plant Mol Biol. 2005 Jul;58(5):643-58. doi: 10.1007/s11103-005-6803-x. Plant Mol Biol. 2005. PMID: 16158240
Cited by
-
Development and selection of stably expressed reference genes for expression normalization in Ribes odoratum under drought stress.Sci Rep. 2025 Aug 9;15(1):29214. doi: 10.1038/s41598-025-12051-1. Sci Rep. 2025. PMID: 40783412 Free PMC article.
References
-
- Baneyx F (1999) Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol 10(5):411–421. 10.1016/s0958-1669(99)00003-8 - PubMed
-
- Bowie JU (2001) Stabilizing membrane proteins. Curr Opin Struct Biol 11(4):397–402. 10.1016/s0959-440x(00)00223-2 - PubMed
-
- Brauer L, Brandt W, Wessjohann LA (2004) Modeling the E. coli 4-hydroxybenzoic acid oligoprenyltransferase (ubiA transferase) and characterization of potential active sites. J Mol Model 10(5–6):317–27. 10.1007/s00894-004-0197-6 - PubMed
-
- Brauer L, Brandt W, Schulze D, Zakharova S, Wessjohann L (2008) A structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. Chembiochem 9(6):982–92. 10.1002/cbic.200700575 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

