Prophage proteins alter long noncoding RNA and DNA of developing sperm to induce a paternal-effect lethality
- PMID: 38452081
- PMCID: PMC11187695
- DOI: 10.1126/science.adk9469
Prophage proteins alter long noncoding RNA and DNA of developing sperm to induce a paternal-effect lethality
Abstract
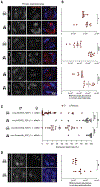
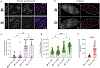
The extent to which prophage proteins interact with eukaryotic macromolecules is largely unknown. In this work, we show that cytoplasmic incompatibility factor A (CifA) and B (CifB) proteins, encoded by prophage WO of the endosymbiont Wolbachia, alter long noncoding RNA (lncRNA) and DNA during Drosophila sperm development to establish a paternal-effect embryonic lethality known as cytoplasmic incompatibility (CI). CifA is a ribonuclease (RNase) that depletes a spermatocyte lncRNA important for the histone-to-protamine transition of spermiogenesis. Both CifA and CifB are deoxyribonucleases (DNases) that elevate DNA damage in late spermiogenesis. lncRNA knockdown enhances CI, and mutagenesis links lncRNA depletion and subsequent sperm chromatin integrity changes to embryonic DNA damage and CI. Hence, prophage proteins interact with eukaryotic macromolecules during gametogenesis to create a symbiosis that is fundamental to insect evolution and vector control.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

