A maternal-effect Padi6 variant causes nuclear and cytoplasmic abnormalities in oocytes, as well as failure of epigenetic reprogramming and zygotic genome activation in embryos
- PMID: 38453481
- PMCID: PMC10982689
- DOI: 10.1101/gad.351238.123
A maternal-effect Padi6 variant causes nuclear and cytoplasmic abnormalities in oocytes, as well as failure of epigenetic reprogramming and zygotic genome activation in embryos
Abstract
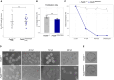
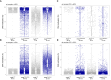
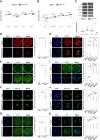
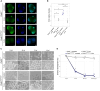
Maternal inactivation of genes encoding components of the subcortical maternal complex (SCMC) and its associated member, PADI6, generally results in early embryo lethality. In humans, SCMC gene variants were found in the healthy mothers of children affected by multilocus imprinting disturbances (MLID). However, how the SCMC controls the DNA methylation required to regulate imprinting remains poorly defined. We generated a mouse line carrying a Padi6 missense variant that was identified in a family with Beckwith-Wiedemann syndrome and MLID. If homozygous in female mice, this variant resulted in interruption of embryo development at the two-cell stage. Single-cell multiomic analyses demonstrated defective maturation of Padi6 mutant oocytes and incomplete DNA demethylation, down-regulation of zygotic genome activation (ZGA) genes, up-regulation of maternal decay genes, and developmental delay in two-cell embryos developing from Padi6 mutant oocytes but little effect on genomic imprinting. Western blotting and immunofluorescence analyses showed reduced levels of UHRF1 in oocytes and abnormal localization of DNMT1 and UHRF1 in both oocytes and zygotes. Treatment with 5-azacytidine reverted DNA hypermethylation but did not rescue the developmental arrest of mutant embryos. Taken together, this study demonstrates that PADI6 controls both nuclear and cytoplasmic oocyte processes that are necessary for preimplantation epigenetic reprogramming and ZGA.
Keywords: DNMT1 localization; Padi6; epigenetic reprogramming; imprinting disorders; maternal-effect genes; multilocus imprinting disturbance; subcortical maternal complex; zygotic genome activation.
© 2024 Giaccari et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Anvar Z, Chakchouk I, Sharif M, Mahadevan S, Nasiotis ET, Su L, Liu Z, Wan YW, Van den Veyver IB. 2023. Loss of the maternal effect gene Nlrp2 alters the transcriptome of ovulated mouse oocytes and impacts expression of histone demethylase KDM1B. Reprod Sci 30: 2780–2793. 10.1007/s43032-023-01218-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
