Targeting metastasis-initiating cancer stem cells in gastric cancer with leukaemia inhibitory factor
- PMID: 38453889
- PMCID: PMC10920825
- DOI: 10.1038/s41420-024-01839-1
Targeting metastasis-initiating cancer stem cells in gastric cancer with leukaemia inhibitory factor
Abstract
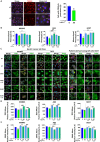
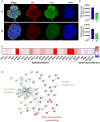
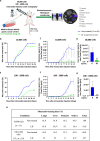
Gastric cancer's (GC) bad prognosis is usually associated with metastatic spread. Invasive cancer stem cells (CSC) are considered to be the seed of GC metastasis and not all CSCs are able to initiate metastasis. Targeting these aggressive metastasis-initiating CSC (MIC) is thus vital. Leukaemia inhibitory factor (LIF) is hereby used to target Hippo pathway oncogenic members, found to be induced in GC and associated with CSC features. LIF-treated GC cell lines, patient-derived xenograft (PDX) cells and/or CSC tumourspheres underwent transcriptomics, laser microdissection-associated proteomics, 2D and 3D invasion assays and in vivo xenograft in mice blood circulation. LIFR expression was analysed on tissue microarrays from GC patients and in silico from public databases. LIF-treated cells, especially CSC, presented decreased epithelial to mesenchymal transition (EMT) phenotype and invasion capacity in vitro, and lower metastasis initiation ability in vivo. These effects involved both the Hippo and Jak/Stat pathways. Finally, GC's high LIFR expression was associated with better clinical outcomes in patients. LIF treatment could thus represent a targeted anti-CSC strategy to fight against metastatic GC, and LIFR detection in primary tumours could constitute a potential new prognosis marker in this disease.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/caac.21660. Epub 4 Feb 2021. - PubMed
-
- Knight WR, Allum WH. Gastric tumours. Medicine. 2019;47:309–13. doi: 10.1016/j.mpmed.2019.02.002. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

