RHBDL4-triggered downregulation of COPII adaptor protein TMED7 suppresses TLR4-mediated inflammatory signaling
- PMID: 38453906
- PMCID: PMC10920636
- DOI: 10.1038/s41467-024-45615-2
RHBDL4-triggered downregulation of COPII adaptor protein TMED7 suppresses TLR4-mediated inflammatory signaling
Abstract
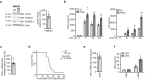
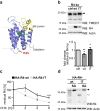
The toll-like receptor 4 (TLR4) is a central regulator of innate immunity that primarily recognizes bacterial lipopolysaccharide cell wall constituents to trigger cytokine secretion. We identify the intramembrane protease RHBDL4 as a negative regulator of TLR4 signaling. We show that RHBDL4 triggers degradation of TLR4's trafficking factor TMED7. This counteracts TLR4 transport to the cell surface. Notably, TLR4 activation mediates transcriptional upregulation of RHBDL4 thereby inducing a negative feedback loop to reduce TLR4 trafficking to the plasma membrane. This secretory cargo tuning mechanism prevents the over-activation of TLR4-dependent signaling in an in vitro Mycobacterium tuberculosis macrophage infection model and consequently alleviates septic shock in a mouse model. A hypomorphic RHBDL4 mutation linked to Kawasaki syndrome, an ill-defined inflammatory disorder in children, further supports the pathophysiological relevance of our findings. In this work, we identify an RHBDL4-mediated axis that acts as a rheostat to prevent over-activation of the TLR4 pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



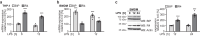

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

