SAM68 directs STING signaling to apoptosis in macrophages
- PMID: 38454028
- PMCID: PMC10920828
- DOI: 10.1038/s42003-024-05969-1
SAM68 directs STING signaling to apoptosis in macrophages
Abstract
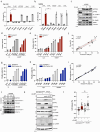
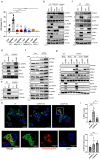
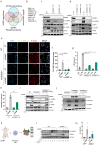
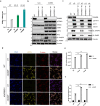
DNA is a danger signal sensed by cGAS to engage signaling through STING to activate innate immune functions. The best-studied downstream responses to STING activation include expression of type I interferon and inflammatory genes, but STING also activates other pathways, including apoptosis. Here, we report that STING-dependent induction of apoptosis in macrophages occurs through the intrinsic mitochondrial pathway and is mediated via IRF3 but acts independently of gene transcription. By intersecting four mass spectrometry datasets, we identify SAM68 as crucial for the induction of apoptosis downstream of STING activation. SAM68 is essential for the full activation of apoptosis. Still, it is not required for STING-mediated activation of IFN expression or activation of NF-κB. Mechanistic studies reveal that protein trafficking is required and involves SAM68 recruitment to STING upon activation, with the two proteins associating at the Golgi or a post-Golgi compartment. Collectively, our work identifies SAM68 as a STING-interacting protein enabling induction of apoptosis through this DNA-activated innate immune pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

