Loss of WIPI4 in neurodegeneration causes autophagy-independent ferroptosis
- PMID: 38454050
- PMCID: PMC11021183
- DOI: 10.1038/s41556-024-01373-3
Loss of WIPI4 in neurodegeneration causes autophagy-independent ferroptosis
Abstract
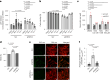
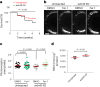
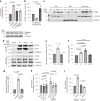
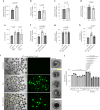
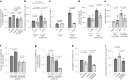
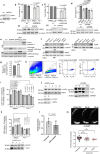
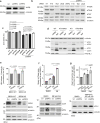
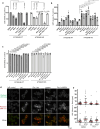
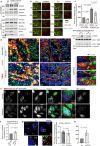
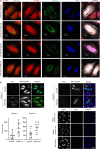
β-Propeller protein-associated neurodegeneration (BPAN) is a rare X-linked dominant disease, one of several conditions that manifest with neurodegeneration and brain iron accumulation. Mutations in the WD repeat domain 45 (WDR45) gene encoding WIPI4 lead to loss of function in BPAN but the cellular mechanisms of how these trigger pathology are unclear. The prevailing view in the literature is that BPAN is simply the consequence of autophagy deficiency given that WIPI4 functions in this degradation pathway. However, our data indicate that WIPI4 depletion causes ferroptosis-a type of cell death induced by lipid peroxidation-via an autophagy-independent mechanism, as demonstrated both in cell culture and in zebrafish. WIPI4 depletion increases ATG2A localization at endoplasmic reticulum-mitochondrial contact sites, which enhances phosphatidylserine import into mitochondria. This results in increased mitochondrial synthesis of phosphatidylethanolamine, a major lipid prone to peroxidation, thus enabling ferroptosis. This mechanism has minimal overlap with classical ferroptosis stimuli but provides insights into the causes of neurodegeneration in BPAN and may provide clues for therapeutic strategies.
© 2024. The Author(s).
Conflict of interest statement
D.C.R. is a consultant for Drishti Discoveries, PAQ Therapeutics, MindRank AI, Retro Biosciences and Nido Biosciences. All other authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

