Identification of genetic suppressors for a BSCL2 lipodystrophy pathogenic variant in Caenorhabditis elegans
- PMID: 38454882
- PMCID: PMC11051982
- DOI: 10.1242/dmm.050524
Identification of genetic suppressors for a BSCL2 lipodystrophy pathogenic variant in Caenorhabditis elegans
Abstract
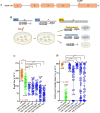
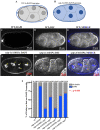
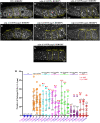
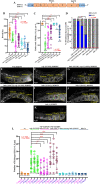
Seipin (BSCL2), a conserved endoplasmic reticulum protein, plays a critical role in lipid droplet (LD) biogenesis and in regulating LD morphology, pathogenic variants of which are associated with Berardinelli-Seip congenital generalized lipodystrophy type 2 (BSCL2). To model BSCL2 disease, we generated an orthologous BSCL2 variant, seip-1(A185P), in Caenorhabditis elegans. In this study, we conducted an unbiased chemical mutagenesis screen to identify genetic suppressors that restore embryonic viability in the seip-1(A185P) mutant background. A total of five suppressor lines were isolated and recovered from the screen. The defective phenotypes of seip-1(A185P), including embryonic lethality and impaired eggshell formation, were significantly suppressed in each suppressor line. Two of the five suppressor lines also alleviated the enlarged LDs in the oocytes. We then mapped a suppressor candidate gene, lmbr-1, which is an ortholog of human limb development membrane protein 1 (LMBR1). The CRISPR/Cas9 edited lmbr-1 suppressor alleles, lmbr-1(S647F) and lmbr-1(P314L), both significantly suppressed embryonic lethality and defective eggshell formation in the seip-1(A185P) background. The newly identified suppressor lines offer valuable insights into potential genetic interactors and pathways that may regulate seipin in the lipodystrophy model.
Keywords: Caenorhabditis elegans; CRISPR/Cas9; Genetic suppressor; LMBR-1; Lipid droplet; Seipin.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
-
- Burns, A. R., Kwok, T. C., Howard, A., Houston, E., Johanson, K., Chan, A., Cutler, S. R., McCourt, P. and Roy, P. J. (2006). High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat. Protoc. 1, 1906-1914. 10.1038/nprot.2006.283 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

