Comprehensive analysis of lactate-related gene profiles and immune characteristics in lupus nephritis
- PMID: 38455045
- PMCID: PMC10917958
- DOI: 10.3389/fimmu.2024.1329009
Comprehensive analysis of lactate-related gene profiles and immune characteristics in lupus nephritis
Abstract
Objectives: The most frequent cause of kidney damage in systemic lupus erythematosus (SLE) is lupus nephritis (LN), which is also a significant risk factor for morbidity and mortality. Lactate metabolism and protein lactylation might be related to the development of LN. However, there is still a lack of relative research to prove the hypothesis. Hence, this study was conducted to screen the lactate-related biomarkers for LN and analyze the underlying mechanism.
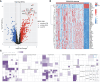
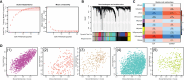
Methods: To identify differentially expressed genes (DEGs) in the training set (GSE32591, GSE127797), we conducted a differential expression analysis (LN samples versus normal samples). Then, module genes were mined using WGCNA concerning LN. The overlapping of DEGs, critical module genes, and lactate-related genes (LRGs) was used to create the lactate-related differentially expressed genes (LR-DEGs). By using a machine-learning algorithm, ROC, and expression levels, biomarkers were discovered. We also carried out an immune infiltration study based on biomarkers and GSEA.
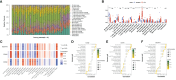
Results: A sum of 1259 DEGs was obtained between LN and normal groups. Then, 3800 module genes in reference to LN were procured. 19 LR-DEGs were screened out by the intersection of DEGs, key module genes, and LRGs. Moreover, 8 pivotal genes were acquired via two machine-learning algorithms. Subsequently, 3 biomarkers related to lactate metabolism were obtained, including COQ2, COQ4, and NDUFV1. And these three biomarkers were enriched in pathways 'antigen processing and presentation' and 'NOD-like receptor signaling pathway'. We found that Macrophages M0 and T cells regulatory (Tregs) were associated with these three biomarkers as well.
Conclusion: Overall, the results indicated that lactate-related biomarkers COQ2, COQ4, and NDUFV1 were associated with LN, which laid a theoretical foundation for the diagnosis and treatment of LN.
Keywords: GEO; bioinformatics; infiltrating immunocytes; lactate; lupus nephritis.
Copyright © 2024 Sun, Gao, Xiang, Feng, Wang, Xu, Wang and Liang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Téllez Noriega JL, Basso V, Fuentes N, Vivero F. Clinical and immunological factors associated with lupus nephritis in an Argentine patient population: A cross-sectional study. Rev Colomb Reumatol. (2022) 29:249–55. doi: 10.1016/j.rcreue.2021.05.003 - DOI
-
- Nezhad ST, Sepaskhah R. Correlation of clinical and pathological findings in patients with lupus nephritis: a five-year experience in Iran. Saudi J Kidney Dis Transpl. (2008) 19(1):32–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

