Detection of evolutionary conserved and accelerated genomic regions related to adaptation to thermal niches in Anolis lizards
- PMID: 38455144
- PMCID: PMC10920033
- DOI: 10.1002/ece3.11117
Detection of evolutionary conserved and accelerated genomic regions related to adaptation to thermal niches in Anolis lizards
Abstract
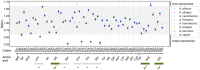
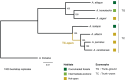
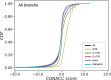
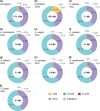
Understanding the genetic basis for adapting to thermal environments is important due to serious effects of global warming on ectothermic species. Various genes associated with thermal adaptation in lizards have been identified mainly focusing on changes in gene expression or the detection of positively selected genes using coding regions. Only a few comprehensive genome-wide analyses have included noncoding regions. This study aimed to identify evolutionarily conserved and accelerated genomic regions using whole genomes of eight Anolis lizard species that have repeatedly adapted to similar thermal environments in multiple lineages. Evolutionarily conserved genomic regions were extracted as regions with overall sequence conservation (regions with fewer base substitutions) across all lineages compared with the neutral model. Genomic regions that underwent accelerated evolution in the lineage of interest were identified as those with more base substitutions in the target branch than in the entire background branch. Conserved elements across all branches were relatively abundant in "intergenic" genomic regions among noncoding regions. Accelerated regions (ARs) of each lineage contained a significantly greater proportion of noncoding RNA genes than the entire multiple alignment. Common genes containing ARs within 5 kb of their vicinity in lineages with similar thermal habitats were identified. Many genes associated with circadian rhythms and behavior were found in hot-open and cool-shaded habitat lineages. These genes might play a role in contributing to thermal adaptation and assist future studies examining the function of genes involved in thermal adaptation via genome editing.
Keywords: Anolis; accelerated evolution; comparative genomics; reptiles; thermal adaptation.
© 2024 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Differentially expressed genes associated with adaptation to different thermal environments in three sympatric Cuban Anolis lizards.Mol Ecol. 2016 May;25(10):2273-85. doi: 10.1111/mec.13625. Epub 2016 May 14. Mol Ecol. 2016. PMID: 27027506
-
Draft genome of six Cuban Anolis lizards and insights into genetic changes during their diversification.BMC Ecol Evol. 2022 Nov 4;22(1):129. doi: 10.1186/s12862-022-02086-7. BMC Ecol Evol. 2022. PMID: 36333669 Free PMC article.
-
A candidate multimodal functional genetic network for thermal adaptation.PeerJ. 2014 Sep 30;2:e578. doi: 10.7717/peerj.578. eCollection 2014. PeerJ. 2014. PMID: 25289178 Free PMC article.
-
The genome of the tegu lizard Salvator merianae: combining Illumina, PacBio, and optical mapping data to generate a highly contiguous assembly.Gigascience. 2018 Dec 1;7(12):giy141. doi: 10.1093/gigascience/giy141. Gigascience. 2018. PMID: 30481296 Free PMC article.
-
Evolutionary conservation in noncoding genomic regions.Trends Genet. 2021 Oct;37(10):903-918. doi: 10.1016/j.tig.2021.06.007. Epub 2021 Jul 5. Trends Genet. 2021. PMID: 34238591 Review.
Cited by
-
Repeatability of evolution and genomic predictions of temperature adaptation in seed beetles.Nat Ecol Evol. 2025 Jun;9(6):1061-1074. doi: 10.1038/s41559-025-02716-5. Epub 2025 May 16. Nat Ecol Evol. 2025. PMID: 40379980 Free PMC article.
References
-
- Alföldi, J. , Di Palma, F. , Grabherr, M. , Williams, C. , Kong, L. , Mauceli, E. , Russell, P. , Lowe, C. B. , Glor, R. E. , Jaffe, J. D. , Ray, D. A. , Boissinot, S. , Shedlock, A. M. , Botka, C. , Castoe, T. A. , Colbourne, J. K. , Fujita, M. K. , Moreno, R. G. , ten Hallers, B. F. , … Lindblad‐Toh, K. (2011). The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature, 477(7366), 587–591. 10.1038/nature10390 - DOI - PMC - PubMed
-
- Blanchette, M. , Kent, W. J. , Riemer, C. , Elnitski, L. , Smit, A. F. , Roskin, K. M. , Baertsch, R. , Rosenbloom, K. , Clawson, H. , Green, E. D. , Haussler, D. , & Miller, W. (2004). Aligning multiple genomic sequences with the threaded blockset aligner. Genome Research, 14(4), 708–715. 10.1101/gr.1933104 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources

