Molecular features of gastroenteropancreatic neuroendocrine carcinoma: A comparative analysis with lung neuroendocrine carcinoma and digestive adenocarcinomas
- PMID: 38455367
- PMCID: PMC10915635
- DOI: 10.21147/j.issn.1000-9604.2024.01.09
Molecular features of gastroenteropancreatic neuroendocrine carcinoma: A comparative analysis with lung neuroendocrine carcinoma and digestive adenocarcinomas
Abstract
Objective: There is an ongoing debate about whether the management of gastroenteropancreatic (GEP) neuroendocrine carcinoma (NEC) should follow the guidelines of small-cell lung cancer (SCLC). We aim to identify the genetic differences of GEPNEC and its counterpart.
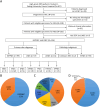
Methods: We recruited GEPNEC patients as the main cohort, with lung NEC and digestive adenocarcinomas as comparative cohorts. All patients undergone next-generation sequencing (NGS). Different gene alterations were compared and analyzed between GEPNEC and lung NEC (LNEC), GEPNEC and adenocarcinoma to yield the remarkable genes.
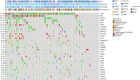
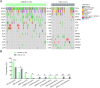
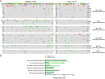
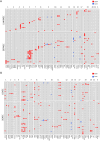
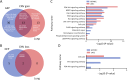
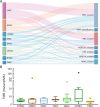
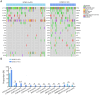
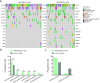
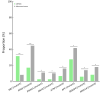
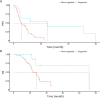
Results: We recruited 257 patients, including 99 GEPNEC, 57 LNEC, and 101 digestive adenocarcinomas. Among the mutations, KRAS, RB1, TERT, IL7R, and CTNNB1 were found to have different gene alterations between GEPNEC and LNEC samples. Specific genes for each site were revealed: gastric NEC ( TERT amplification), colorectal NEC ( KRAS mutation), and bile tract NEC ( ARID1A mutation). The gene disparities between small-cell NEC (SCNEC) and large-cell NEC (LCNEC) were KEAP1 and CDH1. Digestive adenocarcinoma was also compared with GEPNEC and suggested RB1, APC, and KRAS as significant genes. The TP53/ RB1 mutation pattern was associated with first-line effectiveness. Putative targetable genes and biomarkers in GEPNEC were identified in 22.2% of the patients, and they had longer progression-free survival (PFS) upon targetable treatment [12.5 months vs. 3.0 months, HR=0.40 (0.21-0.75), P=0.006].
Conclusions: This work demonstrated striking gene distinctions in GEPNEC compared with LNEC and adenocarcinoma and their clinical utility.
Keywords: Neuroendocrine carcinoma; gastroenteropancreatic; genetic alterations; lung; molecular markers.
Copyright ©2024 Chinese Journal of Cancer Research. All rights reserved.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
