Differential Expression of Noncoding RNAs Revealed Enhancer RNA AC016735.2 as a Potential Pathogenic Marker of Congenital Microtia Patients
- PMID: 38456687
- PMCID: PMC11045553
- DOI: 10.1097/SCS.0000000000010046
Differential Expression of Noncoding RNAs Revealed Enhancer RNA AC016735.2 as a Potential Pathogenic Marker of Congenital Microtia Patients
Abstract
Purpose: Congenital microtia is a complex maxillofacial malformation with various risk factors. This study aimed to find potential pathogenic noncoding RNAs for congenital microtia patients.
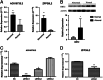
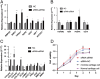
Methods: We collected 3 pairs of residual ear cartilage samples and corresponding normal ear cartilage samples from nonsyndromic congenital microtia patients for microarray experiments. The differentially expressed RNAs were screened, and enrichment analysis and correlation expression analysis were performed to elucidate the function of the differentially expressed genes (DEGs). We further investigated the most significantly differentially expressed long noncoding RNA (lncRNA), AC016735.2, through follow-up analyses including RT-qPCR and Western blotting, to validate its differential expression in residual ear cartilage compared with normal ear cartilage. SiRNA was designed to study the regulatory role of AC016735.2, and cell proliferation experiments were conducted to explore its impact on residual ear chondrocytes.
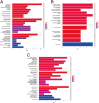
Results: Analysis of the microarray data revealed a total of 1079 differentially expressed RNAs, including 305 mRNAs and x lncRNAs, using a threshold of FC>1.5 and P<0.05 for mRNA, and FC>1.0 and P<0.05 for lncRNA. Enrichment analysis indicated that these genes are mainly involved in extracellular matrix regulation and embryonic development. AC016735.2 showed the highest differential expression among the eRNAs, being upregulated in residual ear cartilage. It acts in cis to regulate the nearby coding gene ZFP36L2, indirectly affecting downstream genes such as BMP4, TWSG1, COL2A1, and COL9A2.
Conclusion: Significant differences were observed in the expression of lncRNAs and mRNAs between residual ear cartilage and normal auricular cartilage tissues in the same genetic background of congenital microtia. These differentially expressed lncRNAs and mRNAs may play crucial roles in the occurrence and development of microtia through pathways associated with extracellular matrix regulation and gastrulation. Particularly, AC016735.2, an eRNA acting in cis, could serve as a potential pathogenic noncoding gene.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of Mutaz B. Habal, MD.
Conflict of interest statement
The authors report no conflicts of interest.
Figures


Similar articles
-
Profiling of Long Non-Coding RNAs in Auricular Cartilage of Patients with Isolated Microtia.Genet Test Mol Biomarkers. 2024 Feb;28(2):50-58. doi: 10.1089/gtmb.2023.0360. Genet Test Mol Biomarkers. 2024. PMID: 38416666
-
Differential expression of long noncoding RNAs in congenital microtia.Gene Expr Patterns. 2017 Nov;25-26:131-141. doi: 10.1016/j.gep.2017.06.007. Epub 2017 Jun 15. Gene Expr Patterns. 2017. PMID: 28625897
-
Protein profile of ear auricle cartilage and the important role of ITGB1/PTK2 in microtia.Int J Pediatr Otorhinolaryngol. 2020 Oct;137:110235. doi: 10.1016/j.ijporl.2020.110235. Epub 2020 Jul 12. Int J Pediatr Otorhinolaryngol. 2020. PMID: 32896350
-
Differential Expression of Long Noncoding RNAs in Murine Myoblasts After Short Hairpin RNA-Mediated Dysferlin Silencing In Vitro: Microarray Profiling.JMIR Bioinform Biotechnol. 2022 Jun 17;3(1):e33186. doi: 10.2196/33186. JMIR Bioinform Biotechnol. 2022. PMID: 38935964 Free PMC article.
-
Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis.Arthritis Rheumatol. 2014 Apr;66(4):969-78. doi: 10.1002/art.38309. Arthritis Rheumatol. 2014. PMID: 24757148
Cited by
-
Systematic Review on Microtia: Current Knowledge and Future Directions.Children (Basel). 2025 Mar 25;12(4):411. doi: 10.3390/children12040411. Children (Basel). 2025. PMID: 40310042 Free PMC article. Review.
References
-
- Forrester MB, Merz RD. Descriptive epidemiology of anotia and microtia, Hawaii, 1986-2002. Congenit Anom (Kyoto) 2005;45:119–124 - PubMed
-
- Suutarla S, Rautio J, Ritvanen A, et al. . Microtia in Finland: comparison of characteristics in different populations. Int J Pediatr Otorhinolaryngol 2007;71:1211–1217 - PubMed
-
- Shaw GM, Carmichael SL, Kaidarova Z, et al. . Epidemiologic characteristics of anotia and microtia in California, 1989-1997. Birth Defects Res A Clin Mol Teratol 2004;70:472–475 - PubMed
-
- Zhang L, Lin L, Song YP, et al. . Differential expression of long noncoding RNAs in congenital microtia. Gene Expr Patterns 2017;25-26:131–141 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

