Large-scale Pan-cancer Cell Line Screening Identifies Actionable and Effective Drug Combinations
- PMID: 38456804
- PMCID: PMC11061612
- DOI: 10.1158/2159-8290.CD-23-0388
Large-scale Pan-cancer Cell Line Screening Identifies Actionable and Effective Drug Combinations
Abstract
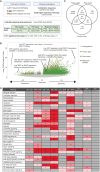
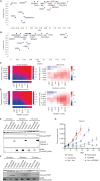
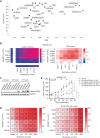
Oncology drug combinations can improve therapeutic responses and increase treatment options for patients. The number of possible combinations is vast and responses can be context-specific. Systematic screens can identify clinically relevant, actionable combinations in defined patient subtypes. We present data for 109 anticancer drug combinations from AstraZeneca's oncology small molecule portfolio screened in 755 pan-cancer cell lines. Combinations were screened in a 7 × 7 concentration matrix, with more than 4 million measurements of sensitivity, producing an exceptionally data-rich resource. We implement a new approach using combination Emax (viability effect) and highest single agent (HSA) to assess combination benefit. We designed a clinical translatability workflow to identify combinations with clearly defined patient populations, rationale for tolerability based on tumor type and combination-specific "emergent" biomarkers, and exposures relevant to clinical doses. We describe three actionable combinations in defined cancer types, confirmed in vitro and in vivo, with a focus on hematologic cancers and apoptotic targets.
Significance: We present the largest cancer drug combination screen published to date with 7 × 7 concentration response matrices for 109 combinations in more than 750 cell lines, complemented by multi-omics predictors of response and identification of "emergent" combination biomarkers. We prioritize hits to optimize clinical translatability, and experimentally validate novel combination hypotheses. This article is featured in Selected Articles from This Issue, p. 695.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Devita VT Jr, Serpick AA, Carbone PP. Combination chemotherapy in the treatment of advanced Hodgkin's disease. Ann Intern Med 1970;73:881–95. - PubMed
-
- DeVita VT Jr. A selective history of the therapy of Hodgkin's disease. Br J Haematol 2003;122:718–27. - PubMed
-
- Boshuizen J, Peeper DS. Rational cancer treatment combinations: an urgent clinical need. Mol Cell 2020;78:1002–18. - PubMed
-
- Morgan P, Brown DG, Lennard S, Anderton MJ, Barrett JC, Eriksson U, et al. . Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nat Rev Drug Discov 2018;17:167–81. - PubMed
-
- Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, et al. . Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat Rev Drug Discov 2014;13:419–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

