The Arabidopsis Information Resource in 2024
- PMID: 38457127
- PMCID: PMC11075553
- DOI: 10.1093/genetics/iyae027
The Arabidopsis Information Resource in 2024
Abstract
Since 1999, The Arabidopsis Information Resource (www.arabidopsis.org) has been curating data about the Arabidopsis thaliana genome. Its primary focus is integrating experimental gene function information from the peer-reviewed literature and codifying it as controlled vocabulary annotations. Our goal is to produce a "gold standard" functional annotation set that reflects the current state of knowledge about the Arabidopsis genome. At the same time, the resource serves as a nexus for community-based collaborations aimed at improving data quality, access, and reuse. For the past decade, our work has been made possible by subscriptions from our global user base. This update covers our ongoing biocuration work, some of our modernization efforts that contribute to the first major infrastructure overhaul since 2011, the introduction of JBrowse2, and the resource's role in community activities such as organizing the structural reannotation of the genome. For gene function assessment, we used gene ontology annotations as a metric to evaluate: (1) what is currently known about Arabidopsis gene function and (2) the set of "unknown" genes. Currently, 74% of the proteome has been annotated to at least one gene ontology term. Of those loci, half have experimental support for at least one of the following aspects: molecular function, biological process, or cellular component. Our work sheds light on the genes for which we have not yet identified any published experimental data and have no functional annotation. Drawing attention to these unknown genes highlights knowledge gaps and potential sources of novel discoveries.
Keywords: biocuration; community resource; model organism database; plant genomics.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflict of interest.
Figures


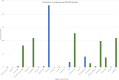




References
-
- Berardini T, Reiser L, Huala E. 2022. TAIR functional annotation data (TAIR_Data_20220331) [Data set]. Zenodo. doi: 10.5281/zenodo.7843882. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

