Transmembrane domain-driven PD-1 dimers mediate T cell inhibition
- PMID: 38457513
- PMCID: PMC11166110
- DOI: 10.1126/sciimmunol.ade6256
Transmembrane domain-driven PD-1 dimers mediate T cell inhibition
Abstract
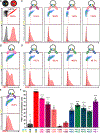
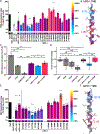
Programmed cell death-1 (PD-1) is a potent immune checkpoint receptor on T lymphocytes. Upon engagement by its ligands, PD-L1 or PD-L2, PD-1 inhibits T cell activation and can promote immune tolerance. Antagonism of PD-1 signaling has proven effective in cancer immunotherapy, and conversely, agonists of the receptor may have a role in treating autoimmune disease. Some immune receptors function as dimers, but PD-1 has been considered monomeric. Here, we show that PD-1 and its ligands form dimers as a consequence of transmembrane domain interactions and that propensity for dimerization correlates with the ability of PD-1 to inhibit immune responses, antitumor immunity, cytotoxic T cell function, and autoimmune tissue destruction. These observations contribute to our understanding of the PD-1 axis and how it can potentially be manipulated for improved treatment of cancer and autoimmune diseases.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

