Effects of L-Leu-L-Leu peptide on growth, proliferation, and apoptosis in broiler intestinal epithelial cells
- PMID: 38457989
- PMCID: PMC11067774
- DOI: 10.1016/j.psj.2024.103582
Effects of L-Leu-L-Leu peptide on growth, proliferation, and apoptosis in broiler intestinal epithelial cells
Abstract
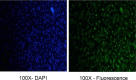
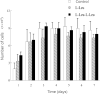
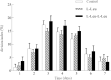
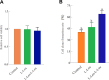
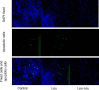
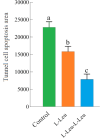
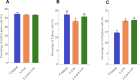
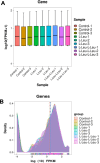
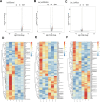
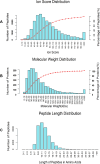
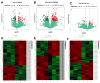
Small peptides are nutrients and bioactive molecules that have dual regulatory effects on nutrition and physiology. They are of great significance for maintaining the intestinal health and production performance of broilers. We here cultured the primary small intestinal epithelial cells (IEC) of chicken in a medium containing L-Leu (Leu) and L-Leu-L-Leu (Leu-Leu) for 24 h. The untreated cells were considered as the control group. The growth, proliferation, and apoptosis of IEC were examined. By combining RNA-seq and label-free sequencing technology, candidate genes, proteins, and pathways related to the growth, proliferation, and apoptosis of IEC were screened. Immunofluorescence detection revealed that the purity of the isolated primary IEC was >90%. The Leu-Leu group significantly promoted IEC growth and proliferation and significantly inhibited IEC apoptosis, and the effect was better than those of the Leu and control groups. Using transcriptome sequencing, four candidate genes, CCL20, IL8L1, IL8, and IL6, were screened in the Leu group, and one candidate gene, IL8, was screened in the Leu-Leu group. Two candidate genes, IL6 and RGN, were screened in the Leu-Leu group compared with the Leu group. Nonquantitative proteomic marker sequencing results revealed that through the screening of candidate proteins and pathways, found one growth-related candidate protein PGM3 and three proliferation-related candidate proteins RPS17, RPS11, and RPL23, and two apoptosis-related candidate proteins GPX4 and PDPK1 were found in the Leu-Leu group compared with Leu group. In short, Leu-Leu could promote IEC growth and proliferation and inhibit IEC apoptosis. On combining transcriptome and proteome sequencing technologies, multiple immune- and energy-related regulatory signal pathways were found to be related to IEC growth, proliferation, and apoptosis. Three candidate genes of IL8, IL6, and RGN were identified, and six candidate proteins of PGM3, RPS17, RPS11, RPL23, GPX4, and PDPK1 were involved in IEC growth, proliferation, and apoptosis. The results provide valuable data for preliminarily elucidating small peptide-mediated IEC regulation pathways, improving the small peptide nutrition theoretical system, and establishing small peptide nutrition regulation technology.
Keywords: apoptosis; candidate gene; candidate protein; cell proliferation; small peptide.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Argiles J.M., Lopezsoriano F.J. Intestinal amino-acid-transport-an overview. Int. J. Biochem. Cell B. 1990;22:931–937. - PubMed
-
- Boza J., Martinez O., Baro L., Suarez M.D., Gil A. Influence of casein and casein hydrolysate diets on nutritional recovery of starved rats. Enter. 1995;19:216–221. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

