An immune cell map of human lung adenocarcinoma development reveals an anti-tumoral role of the Tfh-dependent tertiary lymphoid structure
- PMID: 38458196
- PMCID: PMC10983046
- DOI: 10.1016/j.xcrm.2024.101448
An immune cell map of human lung adenocarcinoma development reveals an anti-tumoral role of the Tfh-dependent tertiary lymphoid structure
Abstract
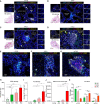
The immune responses during the initiation and invasion stages of human lung adenocarcinoma (LUAD) development are largely unknown. Here, we generated a single-cell RNA sequencing map to decipher the immune dynamics during human LUAD development. We found that T follicular helper (Tfh)-like cells, germinal center B cells, and dysfunctional CD8+ T cells increase during tumor initiation/invasion and form a tertiary lymphoid structure (TLS) inside the tumor. This TLS starts with an aggregation of CD4+ T cells and the generation of CXCL13-expressing Tfh-like cells, followed by an accumulation of B cells, and then forms a CD4+ T and B cell aggregate. TLS and its associated cells are correlated with better patient survival. Inhibiting TLS formation by Tfh or B cell depletion promotes tumor growth in mouse models. The anti-tumoral effect of the Tfh-dependent TLS is mediated through interleukin-21 (IL-21)-IL-21 receptor signaling. Our study establishes an anti-tumoral role of the Tfh-dependent TLS in the development of LUAD.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

