CASZ1 Is Essential for Skin Epidermal Terminal Differentiation
- PMID: 38458428
- PMCID: PMC11344692
- DOI: 10.1016/j.jid.2024.02.014
CASZ1 Is Essential for Skin Epidermal Terminal Differentiation
Abstract
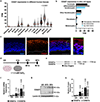
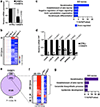
The barrier function of skin epidermis is crucial for our bodies to interface with the environment. Because epidermis continuously turns over throughout the lifetime, this barrier must be actively maintained by regeneration. Although several transcription factors have been established as essential activators in epidermal differentiation, it is unclear whether additional factors remain to be identified. In this study, we show that CASZ1, a multi zinc-finger transcription factor previously characterized in nonepithelial cell types, shows highest expression in skin epidermis. CASZ1 expression is upregulated during epidermal terminal differentiation. In addition, CASZ1 expression is impaired in several skin disorders with impaired barrier function, such as atopic dermatitis, psoriasis, and squamous cell carcinoma. Using transcriptome profiling coupled with RNA interference, we identified 674 differentially expressed genes with CASZ1 knockdown. Downregulated genes account for 91.2% of these differentially expressed genes and were enriched for barrier function. In organotypic epidermal regeneration, CASZ1 knockdown promoted proliferation and strongly impaired multiple terminal differentiation markers. Mechanistically, we found that CASZ1 upregulation in differentiation requires the action of both the master transcription factor, p63, and the histone acetyltransferase, p300. Taken together, our findings identify CASZ1 as an essential activator of epidermal differentiation, paving the way for future studies understanding of CASZ1 roles in skin disease.
Keywords: Barrier function; CASZ1; Differentiation; Keratinocytes; p63.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST
The authors state no conflict of interest.
Figures
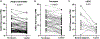


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

