Data-driven energy landscape reveals critical genes in cancer progression
- PMID: 38459043
- PMCID: PMC10923824
- DOI: 10.1038/s41540-024-00354-4
Data-driven energy landscape reveals critical genes in cancer progression
Abstract
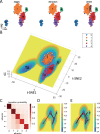
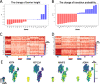
The evolution of cancer is a complex process characterized by stable states and transitions among them. Studying the dynamic evolution of cancer and revealing the mechanisms of cancer progression based on experimental data is an important topic. In this study, we aim to employ a data-driven energy landscape approach to analyze the dynamic evolution of cancer. We take Kidney renal clear cell carcinoma (KIRC) as an example. From the energy landscape, we introduce two quantitative indicators (transition probability and barrier height) to study critical shifts in KIRC cancer evolution, including cancer onset and progression, and identify critical genes involved in these transitions. Our results successfully identify crucial genes that either promote or inhibit these transition processes in KIRC. We also conduct a comprehensive biological function analysis on these genes, validating the accuracy and reliability of our predictions. This work has implications for discovering new biomarkers, drug targets, and cancer treatment strategies in KIRC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Cuproptosis status affects treatment options about immunotherapy and targeted therapy for patients with kidney renal clear cell carcinoma.Front Immunol. 2022 Aug 19;13:954440. doi: 10.3389/fimmu.2022.954440. eCollection 2022. Front Immunol. 2022. PMID: 36059510 Free PMC article.
-
Supervised Learning and Multi-Omics Integration Reveals Clinical Significance of Inner Membrane Mitochondrial Protein (IMMT) in Prognostic Prediction, Tumor Immune Microenvironment and Precision Medicine for Kidney Renal Clear Cell Carcinoma.Int J Mol Sci. 2023 May 15;24(10):8807. doi: 10.3390/ijms24108807. Int J Mol Sci. 2023. PMID: 37240154 Free PMC article.
-
Unique protein expression signatures of survival time in kidney renal clear cell carcinoma through a pan-cancer screening.BMC Genomics. 2017 Oct 3;18(Suppl 6):678. doi: 10.1186/s12864-017-4026-6. BMC Genomics. 2017. PMID: 28984208 Free PMC article.
-
Identification and validation of shared gene signature of kidney renal clear cell carcinoma and COVID-19.PeerJ. 2024 Mar 4;12:e16927. doi: 10.7717/peerj.16927. eCollection 2024. PeerJ. 2024. PMID: 38464749 Free PMC article.
-
Describing a landscape we are yet discovering.Adv Stat Anal. 2022;106(3):399-402. doi: 10.1007/s10182-022-00449-5. Epub 2022 Jun 9. Adv Stat Anal. 2022. PMID: 35698579 Free PMC article. Review. No abstract available.
Cited by
-
Novel marker genes and small molecule drugs for radiotherapy resistance in cervical cancer identified based on single-cell multi-omics analysis.Discov Oncol. 2025 May 19;16(1):823. doi: 10.1007/s12672-025-02532-0. Discov Oncol. 2025. PMID: 40389794 Free PMC article.
References
-
- Sardanyés J, et al. Activation of effector immune cells promotes tumor stochastic extinction: A homotopy analysis approach. Appl Math. Comput. 2015;252:484–495.
-
- Itik M, Salamci MU, Banks SP. Optimal control of drug therapy in cancer treatment. Nonlinear Anal.: Theory, Methods Appl. 2009;71:e1473–e1486. doi: 10.1016/j.na.2009.01.214. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

