Spatially-resolved transcriptomics reveal macrophage heterogeneity and prognostic significance in diffuse large B-cell lymphoma
- PMID: 38459052
- PMCID: PMC10923916
- DOI: 10.1038/s41467-024-46220-z
Spatially-resolved transcriptomics reveal macrophage heterogeneity and prognostic significance in diffuse large B-cell lymphoma
Abstract
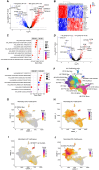
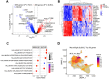
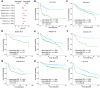
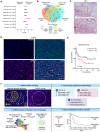
Macrophages are abundant immune cells in the microenvironment of diffuse large B-cell lymphoma (DLBCL). Macrophage estimation by immunohistochemistry shows varying prognostic significance across studies in DLBCL, and does not provide a comprehensive analysis of macrophage subtypes. Here, using digital spatial profiling with whole transcriptome analysis of CD68+ cells, we characterize macrophages in distinct spatial niches of reactive lymphoid tissues (RLTs) and DLBCL. We reveal transcriptomic differences between macrophages within RLTs (light zone /dark zone, germinal center/ interfollicular), and between disease states (RLTs/ DLBCL), which we then use to generate six spatially-derived macrophage signatures (MacroSigs). We proceed to interrogate these MacroSigs in macrophage and DLBCL single-cell RNA-sequencing datasets, and in gene-expression data from multiple DLBCL cohorts. We show that specific MacroSigs are associated with cell-of-origin subtypes and overall survival in DLBCL. This study provides a spatially-resolved whole-transcriptome atlas of macrophages in reactive and malignant lymphoid tissues, showing biological and clinical significance.
© 2024. The Author(s).
Conflict of interest statement
A.D.J. has received consultancy fees from DKSH/Beigene, Roche, Gilead, Turbine Ltd, AstraZeneca, Antengene, Janssen, MSD and IQVIA; and research funding from Janssen and AstraZeneca. The other co-authors have no conflicts of interest to declare.
Figures



References
-
- Coiffier B, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood. 2010;116:2040–2045. doi: 10.1182/blood-2010-03-276246. - DOI - PMC - PubMed
-
- Kotlov N, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov. 2021;11:1468–1489. doi: 10.1158/2159-8290.CD-20-0839. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

