A hybrid modeling framework for generalizable and interpretable predictions of ICU mortality across multiple hospitals
- PMID: 38459085
- PMCID: PMC10923850
- DOI: 10.1038/s41598-024-55577-6
A hybrid modeling framework for generalizable and interpretable predictions of ICU mortality across multiple hospitals
Abstract
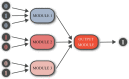
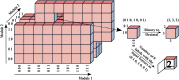
The development of reliable mortality risk stratification models is an active research area in computational healthcare. Mortality risk stratification provides a standard to assist physicians in evaluating a patient's condition or prognosis objectively. Particular interest lies in methods that are transparent to clinical interpretation and that retain predictive power once validated across diverse datasets they were not trained on. This study addresses the challenge of consolidating numerous ICD codes for predictive modeling of ICU mortality, employing a hybrid modeling approach that integrates mechanistic, clinical knowledge with mathematical and machine learning models . A tree-structured network connecting independent modules that carry clinical meaning is implemented for interpretability. Our training strategy utilizes graph-theoretic methods for data analysis, aiming to identify the functions of individual black-box modules within the tree-structured network by harnessing solutions from specific max-cut problems. The trained model is then validated on external datasets from different hospitals, demonstrating successful generalization capabilities, particularly in binary-feature datasets where label assessment involves extrapolation.
Keywords: Generalizability; Hybrid modeling; ICD codes; ICU mortality prediction; Interpretability; Machine learning.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Generalizable calibrated machine learning models for real-time atrial fibrillation risk prediction in ICU patients.Int J Med Inform. 2023 Jul;175:105086. doi: 10.1016/j.ijmedinf.2023.105086. Epub 2023 Apr 26. Int J Med Inform. 2023. PMID: 37148868
-
Noisecut: a python package for noise-tolerant classification of binary data using prior knowledge integration and max-cut solutions.BMC Bioinformatics. 2024 Apr 20;25(1):155. doi: 10.1186/s12859-024-05769-8. BMC Bioinformatics. 2024. PMID: 38641616 Free PMC article.
-
Early Prediction of Cardiac Arrest in the Intensive Care Unit Using Explainable Machine Learning: Retrospective Study.J Med Internet Res. 2024 Sep 17;26:e62890. doi: 10.2196/62890. J Med Internet Res. 2024. PMID: 39288404 Free PMC article.
-
Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: a retrospective study of high-frequency data in electronic patient records.Lancet Digit Health. 2020 Apr;2(4):e179-e191. doi: 10.1016/S2589-7500(20)30018-2. Epub 2020 Mar 12. Lancet Digit Health. 2020. PMID: 33328078
-
Predictive value of machine learning for the risk of acute kidney injury (AKI) in hospital intensive care units (ICU) patients: a systematic review and meta-analysis.PeerJ. 2023 Nov 27;11:e16405. doi: 10.7717/peerj.16405. eCollection 2023. PeerJ. 2023. PMID: 38034868 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

