Sex-specific genetic architecture of blood pressure
- PMID: 38459180
- PMCID: PMC11797078
- DOI: 10.1038/s41591-024-02858-2
Sex-specific genetic architecture of blood pressure
Abstract
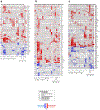
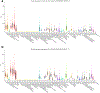
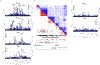
The genetic and genomic basis of sex differences in blood pressure (BP) traits remain unstudied at scale. Here, we conducted sex-stratified and combined-sex genome-wide association studies of BP traits using the UK Biobank resource, identifying 1,346 previously reported and 29 new BP trait-associated loci. Among associated loci, 412 were female-specific (Pfemale ≤ 5 × 10-8; Pmale > 5 × 10-8) and 142 were male-specific (Pmale ≤ 5 × 10-8; Pfemale > 5 × 10-8); these sex-specific loci were enriched for hormone-related transcription factors, in particular, estrogen receptor 1. Analyses of gene-by-sex interactions and sexually dimorphic effects identified four genomic regions, showing female-specific associations with diastolic BP or pulse pressure, including the chromosome 13q34-COL4A1/COL4A2 locus. Notably, female-specific pulse pressure-associated loci exhibited enriched acetylated histone H3 Lys27 modifications in arterial tissues and a female-specific association with fibromuscular dysplasia, a female-biased vascular disease; colocalization signals included Chr13q34: COL4A1/COL4A2, Chr9p21: CDKN2B-AS1 and Chr4q32.1: MAP9 regions. Sex-specific and sex-biased polygenic associations of BP traits were associated with multiple cardiovascular traits. These findings suggest potentially clinically significant and BP sex-specific pleiotropic effects on cardiovascular diseases.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures









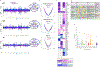


References
METHODS-only references
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

