A multiplex RPA-CRISPR/Cas12a-based POCT technique and its application in human papillomavirus (HPV) typing assay
- PMID: 38459454
- PMCID: PMC10921630
- DOI: 10.1186/s11658-024-00548-y
A multiplex RPA-CRISPR/Cas12a-based POCT technique and its application in human papillomavirus (HPV) typing assay
Erratum in
-
Correction: A multiplex RPA-CRISPR/Cas12a-based POCT technique and its application in human papillomavirus (HPV) typing assay.Cell Mol Biol Lett. 2024 Apr 9;29(1):49. doi: 10.1186/s11658-024-00567-9. Cell Mol Biol Lett. 2024. PMID: 38594613 Free PMC article. No abstract available.
Abstract
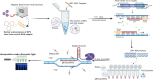
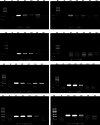
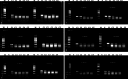
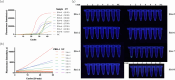
Persistent infection with high-risk human papillomavirus (HR-HPV) is the primary and initiating factor for cervical cancer. With over 200 identified HPV types, including 14 high-risk types that integrate into the host cervical epithelial cell DNA, early determination of HPV infection type is crucial for effective risk stratification and management. Presently, on-site immediate testing during the HPV screening stage, known as Point of Care Testing (POCT), remains immature, severely limiting the scope and scenarios of HPV screening. This study, guided by the genomic sequence patterns of HPV, established a multiplex recombinase polymerase amplification (RPA) technology based on the concept of "universal primers." This approach achieved the multiple amplification of RPA, coupled with the CRISPR/Cas12a system serving as a medium for signal amplification and conversion. The study successfully constructed a POCT combined detection system, denoted as H-MRC12a (HPV-Multiple RPA-CRISPR/Cas12a), and applied it to high-risk HPV typing detection. The system accomplished the typing detection of six high-risk HPV types (16, 18, 31, 33, 35, and 45) can be completed within 40 min, and the entire process, from sample loading to result interpretation, can be accomplished within 45 min, with a detection depth reaching 1 copy/μL for each high-risk type. Validation of the H-MRC12a detection system's reproducibility and specificity was further conducted through QPCR on 34 clinical samples. Additionally, this study explored and optimized the multiplex RPA amplification system and CRISPR system at the molecular mechanism level. Furthermore, the primer design strategy developed in this study offers the potential to enhance the throughput of H-MRC12a detection while ensuring sensitivity, providing a novel research avenue for high-throughput detection in Point-of-Care molecular pathogen studies.
Keywords: CRISPR/Cas12a; HR-HPV; Multiplex RPA; POCT.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









References
-
- World Health Organization . Global strategy to accelerate the elimination of cervical cancer as a public health problem. Geneva: World Health Organization; 2020.
-
- World Health Organization . WHO guideline for screening and treatment of cervical pre-cancer lesions for cervical cancer prevention. Geneva: World Health Organization; 2021. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

