Activation of the PI3K/AKT signaling pathway by ARNTL2 enhances cellular glycolysis and sensitizes pancreatic adenocarcinoma to erlotinib
- PMID: 38459558
- PMCID: PMC10921723
- DOI: 10.1186/s12943-024-01965-5
Activation of the PI3K/AKT signaling pathway by ARNTL2 enhances cellular glycolysis and sensitizes pancreatic adenocarcinoma to erlotinib
Abstract
Background: Pancreatic adenocarcinoma (PC) is an aggressive malignancy with limited treatment options. The poor prognosis primarily stems from late-stage diagnosis and when the disease has become therapeutically challenging. There is an urgent need to identify specific biomarkers for cancer subtyping and early detection to enhance both morbidity and mortality outcomes. The addition of the EGFR tyrosine kinase inhibitor (TKI), erlotinib, to gemcitabine chemotherapy for the first-line treatment of patients with advanced pancreatic cancer slightly improved outcomes. However, restricted clinical benefits may be linked to the absence of well-characterized criteria for stratification and dependable biomarkers for the prediction of treatment effectiveness.
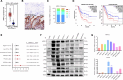
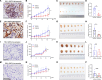
Methods and results: We examined the levels of various cancer hallmarks and identified glycolysis as the primary risk factor for overall survival in PC. Subsequently, we developed a glycolysis-related score (GRS) model to accurately distinguish PC patients with high GRS. Through in silico screening of 4398 compounds, we discovered that erlotinib had the strongest therapeutic benefits for high-GRS PC patients. Furthermore, we identified ARNTL2 as a novel prognostic biomarker and a predictive factor for erlotinib treatment responsiveness in patients with PC. Inhibition of ARNTL2 expression reduced the therapeutic efficacy, whereas increased expression of ARNTL2 improved PC cell sensitivity to erlotinib. Validation in vivo using patient-derived xenografts (PDX-PC) with varying ARNTL2 expression levels demonstrated that erlotinib monotherapy effectively halted tumor progression in PDX-PC models with high ARNTL2 expression. In contrast, PDX-PC models lacking ARNTL2 did not respond favorably to erlotinib treatment. Mechanistically, we demonstrated that the ARNTL2/E2F1 axis-mediated cellular glycolysis sensitizes PC cells to erlotinib treatment by activating the PI3K/AKT signaling pathway.
Conclusions: Our investigations have identified ARNTL2 as a novel prognostic biomarker and predictive indicator of sensitivity. These results will help to identify erlotinib-responsive cases of PC and improve treatment outcomes. These findings contribute to the advancement of precision oncology, enabling more accurate and targeted therapeutic interventions.
Keywords: ARNTL2; Erlotinib; Pancreatic adenocarcinoma; Prognosis; Targeted therapy, precision oncology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
- 81972280/National Natural Science Foundation of China
- 82103087/National Natural Science Foundation of China
- 82171824/National Natural Science Foundation of China
- 82272906/National Natural Science Foundation of China
- 20YF1446400/Shanghai Sailing Program
- 21JC1404300/Scientific and Technological Innovation Project of Science and Technology Commission of Shanghai Municipality
- 2019CXJQ03/Innovation Group Project of Shanghai Municipal Health Commission
- 2018ZHYL0223/Shanghai Municipal Commission of Health and Family Planning Grant
- 20161312/Shanghai Municipal Education Commission-Gao Feng Clinical Medicine Grant Support
- Oncology/Shanghai Key Clinical Specialty
- No. SHDC2020CR1035B/Shanghai Leading Talents Project; Clinical Research Plan of SHDC
- 2019YFC1315900/National Key R&D Program of China
- Y- 2019AZZD-0513/Project from CSCO Clinical Oncology Research Foundation
- PWRd2022-02/Academic Leaders Training Program of Pudong Health Bureau of Shanghai
- SHSMU-ZDCX20210802/the Innovative Research Team of High-Level Local Universities in Shanghai
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

