Alternative splicing expands the clinical spectrum of NDUFS6-related mitochondrial disorders
- PMID: 38459834
- PMCID: PMC11180951
- DOI: 10.1016/j.gim.2024.101117
Alternative splicing expands the clinical spectrum of NDUFS6-related mitochondrial disorders
Abstract
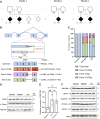
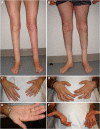
Purpose: We describe 3 families with Charcot-Marie-Tooth neuropathy (CMT), harboring a homozygous NDUFS6 NM_004553.6:c.309+5G>A variant previously linked to fatal Leigh syndrome. We aimed to characterize clinically and molecularly the newly identified patients and understand the mechanism underlying their milder phenotype.
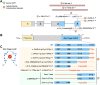
Methods: The patients underwent extensive clinical examinations. Exome sequencing was done in 4 affected individuals. The functional effect of the c.309+5G>A variant was investigated in patient-derived EBV-transformed lymphoblasts at the complementary DNA, protein, and mitochondrial level. Alternative splicing was evaluated using complementary DNA long-read sequencing.
Results: All patients presented with early-onset, slowly progressive axonal CMT, and nystagmus; some exhibited additional central nervous system symptoms. The c.309+5G>A substitution caused the expression of aberrantly spliced transcripts and negligible levels of the canonical transcript. Immunoblotting showed reduced levels of mutant isoforms. No detectable defects in mitochondrial complex stability or bioenergetics were found.
Conclusion: We expand the clinical spectrum of NDUFS6-related mitochondrial disorders to include axonal CMT, emphasizing the clinical and pathophysiologic overlap between these 2 clinical entities. This work demonstrates the critical role that alternative splicing may play in modulating the severity of a genetic disorder, emphasizing the need for careful consideration when interpreting splice variants and their implications on disease prognosis.
Keywords: Charcot-Marie-Tooth; Mitochondrial disorders; NDUFS6; Peripheral neuropathy; Splicing.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

