Ecological Predictors of Organelle Genome Evolution: Phylogenetic Correlations with Taxonomically Broad, Sparse, Unsystematized Data
- PMID: 38459872
- PMCID: PMC11282362
- DOI: 10.1093/sysbio/syae009
Ecological Predictors of Organelle Genome Evolution: Phylogenetic Correlations with Taxonomically Broad, Sparse, Unsystematized Data
Abstract
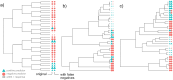
Comparative analysis of variables across phylogenetically linked observations can reveal mechanisms and insights in evolutionary biology. As the taxonomic breadth of the sample of interest increases, challenges of data sparsity, poor phylogenetic resolution, and complicated evolutionary dynamics emerge. Here, we investigate a cross-eukaryotic question where all these problems exist: which organismal ecology features are correlated with gene retention in mitochondrial and chloroplast DNA (organelle DNA or oDNA). Through a wide palette of synthetic control studies, we first characterize the specificity and sensitivity of a collection of parametric and non-parametric phylogenetic comparative approaches to identify relationships in the face of such sparse and awkward datasets. This analysis is not directly focused on oDNA, and so provides generalizable insights into comparative approaches with challenging data. We then combine and curate ecological data coupled to oDNA genome information across eukaryotes, including a new semi-automated approach for gathering data on organismal traits from less systematized open-access resources including encyclopedia articles on species and taxa. The curation process also involved resolving several issues with existing datasets, including enforcing the clade-specificity of several ecological features and fixing incorrect annotations. Combining this unique dataset with our benchmarked comparative approaches, we confirm support for several known links between organismal ecology and organelle gene retention, identify several previously unidentified relationships constituting possible ecological contributors to oDNA genome evolution, and provide support for a recently hypothesized link between environmental demand and oDNA retention. We, with caution, discuss the implications of these findings for organelle evolution and of this pipeline for broad comparative analyses in other fields.
Keywords: Comparative methods; ecology; mtDNA; organelle evolution; phylogenetic generalized linear model; phylogenetic linear model; ptDNA.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society of Systematic Biologists.
Figures



References
-
- Auguie B., Antonov A. 2017. gridExtra: miscellaneous functions for “Grid” graphics. R Package Version 2.3. Computer Software. Available from: https://cran.r-project.org/web/packages/gridExtra/.
-
- Bates D., Mächler M., Bolker B., Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67(October):1–48. doi: 10.18637/jss.v067.i01 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

