BIN1K358R suppresses glial response to plaques in mouse model of Alzheimer's disease
- PMID: 38460121
- PMCID: PMC11032570
- DOI: 10.1002/alz.13767
BIN1K358R suppresses glial response to plaques in mouse model of Alzheimer's disease
Abstract
Introduction: The BIN1 coding variant rs138047593 (K358R) is linked to Late-Onset Alzheimer's Disease (LOAD) via targeted exome sequencing.
Methods: To elucidate the functional consequences of this rare coding variant on brain amyloidosis and neuroinflammation, we generated BIN1K358R knock-in mice using CRISPR/Cas9 technology. These mice were subsequently bred with 5xFAD transgenic mice, which serve as a model for Alzheimer's pathology.
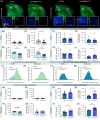
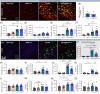
Results: The presence of the BIN1K358R variant leads to increased cerebral amyloid deposition, with a dampened response of astrocytes and oligodendrocytes, but not microglia, at both the cellular and transcriptional levels. This correlates with decreased neurofilament light chain in both plasma and brain tissue. Synaptic densities are significantly increased in both wild-type and 5xFAD backgrounds homozygous for the BIN1K358R variant.
Discussion: The BIN1 K358R variant modulates amyloid pathology in 5xFAD mice, attenuates the astrocytic and oligodendrocytic responses to amyloid plaques, decreases damage markers, and elevates synaptic densities.
Highlights: BIN1 rs138047593 (K358R) coding variant is associated with increased risk of LOAD. BIN1 K358R variant increases amyloid plaque load in 12-month-old 5xFAD mice. BIN1 K358R variant dampens astrocytic and oligodendrocytic response to plaques. BIN1 K358R variant decreases neuronal damage in 5xFAD mice. BIN1 K358R upregulates synaptic densities and modulates synaptic transmission.
Keywords: Alzheimer's disease; BIN1 K358R; MODEL‐AD; astrocytes; inflammation; oligodendrocytes.
© 2024 The Authors. Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Author disclosures are available in the supporting information.
Figures





References
-
- Luukkainen L, Helisalmi S, Kytovuori L, et al. Mutation analysis of the genes linked to early onset Alzheimer's disease and frontotemporal lobar degeneration. J Alzheimers Dis. 2019;69:775‐782. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

