Re-analysis of an outbreak of Shiga toxin-producing Escherichia coli O157:H7 associated with raw drinking milk using Nanopore sequencing
- PMID: 38461188
- PMCID: PMC10925052
- DOI: 10.1038/s41598-024-54662-0
Re-analysis of an outbreak of Shiga toxin-producing Escherichia coli O157:H7 associated with raw drinking milk using Nanopore sequencing
Abstract
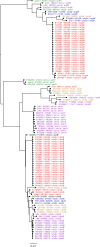
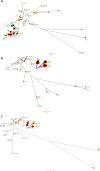
The aim of this study was to compare Illumina and Oxford Nanopore Technology (ONT) sequencing data to quantify genetic variation to assess within-outbreak strain relatedness and characterise microevolutionary events in the accessory genomes of a cluster of 23 genetically and epidemiologically linked isolates related to an outbreak of Shiga toxin-producing Escherichia coli O157:H7 caused by the consumption of raw drinking milk. There were seven discrepant variants called between the two technologies, five were false-negative or false-positive variants in the Illumina data and two were false-negative calls in ONT data. After masking horizontally acquired sequences such as prophages, analysis of both short and long-read sequences revealed the 20 isolates linked to the outbreak in 2017 had a maximum SNP distance of one SNP between each other, and a maximum of five SNPs when including three additional strains identified in 2019. Analysis of the ONT data revealed a 47 kbp deletion event in a terminal compound prophage within one sample relative to the remaining samples, and a 0.65 Mbp large chromosomal rearrangement (inversion), within one sample relative to the remaining samples. Furthermore, we detected two bacteriophages encoding the highly pathogenic Shiga toxin (Stx) subtype, Stx2a. One was typical of Stx2a-phage in this sub-lineage (Ic), the other was atypical and inserted into a site usually occupied by Stx2c-encoding phage. Finally, we observed an increase in the size of the pO157 IncFIB plasmid (1.6 kbp) in isolates from 2019 compared to those from 2017, due to the duplication of insertion elements within the plasmids from the more recently isolated strains. The ability to characterize the accessory genome in this way is the first step to understanding the significance of these microevolutionary events and their impact on the genome plasticity and virulence between strains of this zoonotic, foodborne pathogen.
Keywords: Foodborne outbreak; Genomic epidemiology; Illumina; Nanopore; Prophage comparison; STEC O157:H7; WGS.
© 2024. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Analysis of a small outbreak of Shiga toxin-producing Escherichia coli O157:H7 using long-read sequencing.Microb Genom. 2021 Mar;7(3):mgen000545. doi: 10.1099/mgen.0.000545. Epub 2021 Mar 8. Microb Genom. 2021. PMID: 33683192 Free PMC article.
-
Comparison of Shiga toxin-encoding bacteriophages in highly pathogenic strains of Shiga toxin-producing Escherichia coli O157:H7 in the UK.Microb Genom. 2020 Mar;6(3):e000334. doi: 10.1099/mgen.0.000334. Epub 2020 Feb 24. Microb Genom. 2020. PMID: 32100710 Free PMC article.
-
Differential induction of Shiga toxin in environmental Escherichia coli O145:H28 strains carrying the same genotype as the outbreak strains.Int J Food Microbiol. 2021 Feb 2;339:109029. doi: 10.1016/j.ijfoodmicro.2020.109029. Epub 2020 Dec 23. Int J Food Microbiol. 2021. PMID: 33360585
-
Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain.Int Microbiol. 2011 Sep;14(3):121-41. doi: 10.2436/20.1501.01.142. Int Microbiol. 2011. PMID: 22101411 Review.
-
Methods for the detection and isolation of Shiga toxin-producing Escherichia coli.Symp Ser Soc Appl Microbiol. 2000;(29):133S-143S. doi: 10.1111/j.1365-2672.2000.tb05341.x. Symp Ser Soc Appl Microbiol. 2000. PMID: 10880188 Review.
Cited by
-
Microbiological Analysis Conducted on Raw Milk Collected During Official Sampling in Liguria (North-West Italy) over a Ten-Year Period (2014-2023).Animals (Basel). 2025 Jan 20;15(2):286. doi: 10.3390/ani15020286. Animals (Basel). 2025. PMID: 39858286 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

