Mi-2β promotes immune evasion in melanoma by activating EZH2 methylation
- PMID: 38461299
- PMCID: PMC10924921
- DOI: 10.1038/s41467-024-46422-5
Mi-2β promotes immune evasion in melanoma by activating EZH2 methylation
Abstract
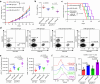
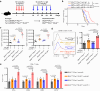
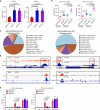
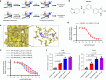
Recent development of new immune checkpoint inhibitors has been particularly successfully in cancer treatment, but still the majority patients fail to benefit. Converting resistant tumors to immunotherapy sensitive will provide a significant improvement in patient outcome. Here we identify Mi-2β as a key melanoma-intrinsic effector regulating the adaptive anti-tumor immune response. Studies in genetically engineered mouse melanoma models indicate that loss of Mi-2β rescues the immune response to immunotherapy in vivo. Mechanistically, ATAC-seq analysis shows that Mi-2β controls the accessibility of IFN-γ-stimulated genes (ISGs). Mi-2β binds to EZH2 and promotes K510 methylation of EZH2, subsequently activating the trimethylation of H3K27 to inhibit the transcription of ISGs. Finally, we develop an Mi-2β-targeted inhibitor, Z36-MP5, which reduces Mi-2β ATPase activity and reactivates ISG transcription. Consequently, Z36-MP5 induces a response to immune checkpoint inhibitors in otherwise resistant melanoma models. Our work provides a potential therapeutic strategy to convert immunotherapy resistant melanomas to sensitive ones.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

