TERRA-LSD1 phase separation promotes R-loop formation for telomere maintenance in ALT cancer cells
- PMID: 38461301
- PMCID: PMC10925046
- DOI: 10.1038/s41467-024-46509-z
TERRA-LSD1 phase separation promotes R-loop formation for telomere maintenance in ALT cancer cells
Abstract
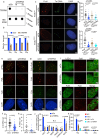
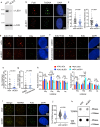
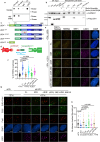
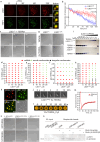
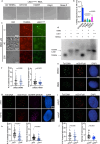
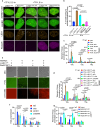
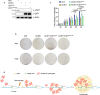
The telomere repeat-containing RNA (TERRA) forms R-loops to promote homology-directed DNA synthesis in the alternative lengthening of telomere (ALT) pathway. Here we report that TERRA contributes to ALT via interacting with the lysine-specific demethylase 1A (LSD1 or KDM1A). We show that LSD1 localizes to ALT telomeres in a TERRA dependent manner and LSD1 function in ALT is largely independent of its demethylase activity. Instead, LSD1 promotes TERRA recruitment to ALT telomeres via RNA binding. In addition, LSD1 and TERRA undergo phase separation, driven by interactions between the RNA binding properties of LSD1 and the G-quadruplex structure of TERRA. Importantly, the formation of TERRA-LSD1 condensates enriches the R-loop stimulating protein Rad51AP1 and increases TERRA-containing R-loops at telomeres. Our findings suggest that LSD1-TERRA phase separation enhances the function of R-loop regulatory molecules for ALT telomere maintenance, providing a mechanism for how the biophysical properties of histone modification enzyme-RNA interactions impact chromatin function.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

