FishSNP: a high quality cross-species SNP database of fishes
- PMID: 38461307
- PMCID: PMC10924876
- DOI: 10.1038/s41597-024-03111-8
FishSNP: a high quality cross-species SNP database of fishes
Abstract
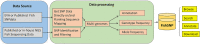
The progress of aquaculture heavily depends on the efficient utilization of diverse genetic resources to enhance production efficiency and maximize profitability. Single nucleotide polymorphisms (SNPs) have been widely used in the study of aquaculture genomics, genetics, and breeding research since they are the most prevalent molecular markers on the genome. Currently, a large number of SNP markers from cultured fish species are scattered in individual studies, making querying complicated and data reuse problematic. We compiled relevant SNP data from literature and public databases to create a fish SNP database, FishSNP ( http://bioinfo.ihb.ac.cn/fishsnp ), and also used a unified analysis pipeline to process raw data that the author of the literature did not perform SNP calling on to obtain SNPs with high reliability. This database presently contains 45,690,243 (45 million) nonredundant SNP data for 13 fish species, with 30,288,958 (30 million) of those being high-quality SNPs. The main function of FishSNP is to search, browse, annotate and download SNPs, which provide researchers various and comprehensive associated information.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Animal-SNPAtlas: a comprehensive SNP database for multiple animals.Nucleic Acids Res. 2023 Jan 6;51(D1):D816-D826. doi: 10.1093/nar/gkac954. Nucleic Acids Res. 2023. PMID: 36300636 Free PMC article.
-
SR4R: An Integrative SNP Resource for Genomic Breeding and Population Research in Rice.Genomics Proteomics Bioinformatics. 2020 Apr;18(2):173-185. doi: 10.1016/j.gpb.2020.03.002. Epub 2020 Jun 30. Genomics Proteomics Bioinformatics. 2020. PMID: 32619768 Free PMC article.
-
Application of DArT seq derived SNP tags for comparative genome analysis in fishes; An alternative pipeline using sequence data from a non-traditional model species, Macquaria ambigua.PLoS One. 2019 Dec 12;14(12):e0226365. doi: 10.1371/journal.pone.0226365. eCollection 2019. PLoS One. 2019. PMID: 31830141 Free PMC article.
-
A SNP-centric database for the investigation of the human genome.BMC Bioinformatics. 2004 Mar 26;5:33. doi: 10.1186/1471-2105-5-33. BMC Bioinformatics. 2004. PMID: 15046636 Free PMC article. Review.
-
SNP web resources and their potential applications in personalized medicine.Curr Drug Metab. 2012 Sep 1;13(7):978-90. doi: 10.2174/138920012802138552. Curr Drug Metab. 2012. PMID: 22591348 Review.
Cited by
-
Chromosome-level genome assembly and annotation of the gynogenetic large-scale loach (Paramisgurnus dabryanus).Sci Data. 2025 Jan 26;12(1):155. doi: 10.1038/s41597-025-04498-8. Sci Data. 2025. PMID: 39865085 Free PMC article.
-
A comparative review of short genetic variant databases across humans and animal species.Brief Bioinform. 2025 Jul 2;26(4):bbaf356. doi: 10.1093/bib/bbaf356. Brief Bioinform. 2025. PMID: 40736747 Free PMC article. Review.
References
-
- Sun Y-L, et al. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus) Aquaculture. 2014;433:19–27. doi: 10.1016/j.aquaculture.2014.05.035. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources