Proteomic analysis of mitochondria associated membranes in renal ischemic reperfusion injury
- PMID: 38461333
- PMCID: PMC10925013
- DOI: 10.1186/s12967-024-05021-0
Proteomic analysis of mitochondria associated membranes in renal ischemic reperfusion injury
Abstract
Background: The mitochondria and endoplasmic reticulum (ER) communicate via contact sites known as mitochondria associated membranes (MAMs). Many important cellular functions such as bioenergetics, mitophagy, apoptosis, and calcium signaling are regulated by MAMs, which are thought to be closely related to ischemic reperfusion injury (IRI). However, there exists a gap in systematic proteomic research addressing the relationship between these cellular processes.
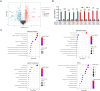
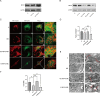
Methods: A 4D label free mass spectrometry-based proteomic analysis of mitochondria associated membranes (MAMs) from the human renal proximal tubular epithelial cell line (HK-2 cells) was conducted under both normal (N) and hypoxia/reperfusion (HR) conditions. Subsequent differential proteins analysis aimed to characterize disease-relevant signaling molecules. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was applied to total proteins and differentially expressed proteins, encompassing Biological Process (BP), Cell Component (CC), Molecular Function (MF), and KEGG pathways. Further, Protein-Protein Interaction Network (PPI) exploration was carried out, leading to the identification of hub genes from differentially expressed proteins. Notably, Mitofusion 2 (MFN2) and BCL2/Adenovirus E1B 19-kDa interacting protein 3(BNIP3) were identified and subsequently validated both in vitro and in vivo. Finally, the impact of MFN2 on MAMs during hypoxia/reoxygenation was explored through regulation of gene expression. Subsequently, a comparative proteomics analysis was conducted between OE-MFN2 and normal HK-2 cells, providing further insights into the underlying mechanisms.
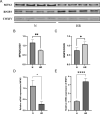
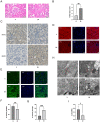
Results: A total of 4489 proteins were identified, with 3531 successfully quantified. GO/KEGG analysis revealed that MAM proteins were primarily associated with mitochondrial function and energy metabolism. Differential analysis between the two groups showed that 688 proteins in HR HK-2 cells exhibited significant changes in expression level with P-value < 0.05 and HR/N > 1.5 or HR/N < 0.66 set as the threshold criteria. Enrichment analysis of differentially expressed proteins unveiled biological processes such as mRNA splicing, apoptosis regulation, and cell division, while molecular functions were predominantly associated with energy metabolic activity. These proteins play key roles in the cellular responses during HR, offering insights into the IRI mechanisms and potential therapeutic targets. The validation of hub genes MFN2 and BNIP3 both in vitro and vivo was consistent with the proteomic findings. MFN2 demonstrated a protective role in maintaining the integrity of mitochondria associated membranes (MAMs) and mitigating mitochondrial damage following hypoxia/reoxygenation injury, this protective effect may be associated with the activation of the PI3K/AKT pathway.
Conclusions: The proteins located in mitochondria associated membranes (MAMs) are implicated in crucial roles during renal ischemic reperfusion injury (IRI), with MFN2 playing a pivotal regulatory role in this context.
Keywords: Ischemic reperfusion injury; Kidney; Mass spectrometry; Mitochondria associated membranes; Proteomics analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Zhao M, Wang Y, Li L, Liu S, Wang C, Yuan Y, Yang G, Chen Y, Cheng J, Lu Y, et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics. 2021;11(4):1845–1863. doi: 10.7150/thno.50905. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

