Genome-wide CRISPR activation screen identifies JADE3 as an antiviral activator of NF-kB-dependent IFITM3 expression
- PMID: 38462163
- PMCID: PMC11001640
- DOI: 10.1016/j.jbc.2024.107153
Genome-wide CRISPR activation screen identifies JADE3 as an antiviral activator of NF-kB-dependent IFITM3 expression
Abstract
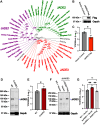
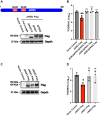
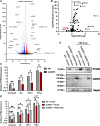
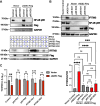
The innate immune system features a web of interacting pathways that require exquisite regulation. To identify novel nodes in this immune landscape, we conducted a gain-of-function, genome-wide CRISPR activation screen with influenza A virus. We identified both appreciated and novel antiviral genes, including Jade family PHD zinc finger 3 (JADE3) a protein involved in directing the histone acetyltransferase histone acetyltransferase binding to ORC1 complex to modify chromatin and regulate transcription. JADE3 is both necessary and sufficient to restrict influenza A virus infection. Our results suggest a distinct function for JADE3 as expression of the closely related paralogs JADE1 and JADE2 does not confer resistance to influenza A virus infection. JADE3 is required for both constitutive and inducible expression of the well-characterized antiviral gene interferon-induced transmembrane protein 3 (IFITM3). Furthermore, we find JADE3 activates the NF-kB signaling pathway, which is required for the promotion of IFITM3 expression by JADE3. Therefore, we propose JADE3 activates an antiviral genetic program involving NF-kB-dependent IFITM3 expression to restrict influenza A virus infection.
Keywords: CRISPR/Cas9 screen; IFITM3; JADE; NF-kB; histone acetylation; inflammation; influenza A virus.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interests with the contents of this article.
Figures




Update of
-
Genome-wide CRISPR activation screen identifies JADE3 as an antiviral activator of NF-kB.bioRxiv [Preprint]. 2023 Sep 29:2023.09.29.560128. doi: 10.1101/2023.09.29.560128. bioRxiv. 2023. Update in: J Biol Chem. 2024 Apr;300(4):107153. doi: 10.1016/j.jbc.2024.107153. PMID: 37808733 Free PMC article. Updated. Preprint.
References
-
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

