Genetic dissection of mutagenic repair and T-DNA capture at CRISPR-induced DNA breaks in Arabidopsis thaliana
- PMID: 38463035
- PMCID: PMC10923293
- DOI: 10.1093/pnasnexus/pgae094
Genetic dissection of mutagenic repair and T-DNA capture at CRISPR-induced DNA breaks in Arabidopsis thaliana
Abstract
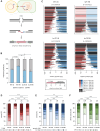
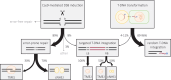
A practical and powerful approach for genome editing in plants is delivery of CRISPR reagents via Agrobacterium tumefaciens transformation. The double-strand break (DSB)-inducing enzyme is expressed from a transferred segment of bacterial DNA, the T-DNA, which upon transformation integrates at random locations into the host genome or is captured at the self-inflicted DSB site. To develop efficient strategies for precise genome editing, it is thus important to define the mechanisms that repair CRISPR-induced DSBs, as well as those that govern random and targeted integration of T-DNA. In this study, we present a detailed and comprehensive genetic analysis of Cas9-induced DSB repair and T-DNA capture in the model plant Arabidopsis thaliana. We found that classical nonhomologous end joining (cNHEJ) and polymerase theta-mediated end joining (TMEJ) are both, and in part redundantly, acting on CRISPR-induced DSBs to produce very different mutational outcomes. We used newly developed CISGUIDE technology to establish that 8% of mutant alleles have captured T-DNA at the induced break site. In addition, we find T-DNA shards within genomic DSB repair sites indicative of frequent temporary interactions during TMEJ. Analysis of thousands of plant genome-T-DNA junctions, followed up by genetic dissection, further reveals that TMEJ is responsible for attaching the 3' end of T-DNA to a CRISPR-induced DSB, while the 5' end can be attached via TMEJ as well as cNHEJ. By identifying the mechanisms that act to connect recombinogenic ends of DNA molecules at chromosomal breaks, and quantifying their contributions, our study supports the development of tailor-made strategies toward predictable engineering of crop plants.
Keywords: Arabidopsis thaliana; DNA repair; T-DNA; classical nonhomologous end joining (cNHEJ); polymerase theta-mediated end joining (TMEJ).
© The Author(s) 2024. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures




Similar articles
-
Division of labor within polymerase theta in repair of CRISPR-induced DNA breaks in Arabidopsis thaliana.PNAS Nexus. 2025 Jun 3;4(6):pgaf183. doi: 10.1093/pnasnexus/pgaf183. eCollection 2025 Jun. PNAS Nexus. 2025. PMID: 40501453 Free PMC article.
-
Profiling Cas9- and Cas12a-induced mutagenesis in Arabidopsis thaliana.Plant J. 2024 Sep;119(6):2706-2717. doi: 10.1111/tpj.16943. Epub 2024 Jul 25. Plant J. 2024. PMID: 39052360
-
Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells.EMBO J. 2017 Dec 15;36(24):3634-3649. doi: 10.15252/embj.201796948. Epub 2017 Oct 27. EMBO J. 2017. PMID: 29079701 Free PMC article.
-
Templated Insertions: A Smoking Gun for Polymerase Theta-Mediated End Joining.Trends Genet. 2019 Sep;35(9):632-644. doi: 10.1016/j.tig.2019.06.001. Epub 2019 Jul 8. Trends Genet. 2019. PMID: 31296341 Review.
-
The biological principles and advanced applications of DSB repair in CRISPR-mediated yeast genome editing.Synth Syst Biotechnol. 2023 Aug 30;8(4):584-596. doi: 10.1016/j.synbio.2023.08.007. eCollection 2023 Dec. Synth Syst Biotechnol. 2023. PMID: 37711546 Free PMC article. Review.
Cited by
-
Manipulating alternative end-joining alters carbon-ion beam-induced genome mutation profiles in Arabidopsis thaliana.DNA Res. 2025 Jul 4;32(4):dsaf014. doi: 10.1093/dnares/dsaf014. DNA Res. 2025. PMID: 40448271 Free PMC article.
-
Context effects on repair of 5'-overhang DNA double-strand breaks induced by Cas12a in Arabidopsis.Plant Direct. 2024 Oct 17;8(10):e70009. doi: 10.1002/pld3.70009. eCollection 2024 Oct. Plant Direct. 2024. PMID: 39421463 Free PMC article.
-
Division of labor within polymerase theta in repair of CRISPR-induced DNA breaks in Arabidopsis thaliana.PNAS Nexus. 2025 Jun 3;4(6):pgaf183. doi: 10.1093/pnasnexus/pgaf183. eCollection 2025 Jun. PNAS Nexus. 2025. PMID: 40501453 Free PMC article.
References
LinkOut - more resources
Full Text Sources

