Genotype-by-environment interactions govern fitness changes associated with adaptive mutations in two-component response systems
- PMID: 38463171
- PMCID: PMC10920338
- DOI: 10.3389/fgene.2024.1349507
Genotype-by-environment interactions govern fitness changes associated with adaptive mutations in two-component response systems
Abstract
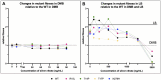
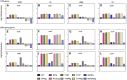
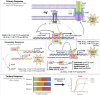
Introduction: Two-component response systems (TCRS) are the main mechanism by which prokaryotes acclimate to changing environments. These systems are composed of a membrane bound histidine kinase (HK) that senses external signals and a response regulator (RR) that activates transcription of response genes. Despite their known role in acclimation, little is known about the role TCRS play in environmental adaptation. Several experimental evolution studies have shown the acquisition of mutations in TCRS during adaptation, therefore here we set out to characterize the adaptive mechanism resulting from these mutations and evaluate whether single nucleotide changes in one gene could induce variable genotype-by-environment (GxE) interactions. Methods: To do this, we assessed fitness changes and differential gene expression for four adaptive mutations in cusS, the gene that encodes the HK CusS, acquired by Escherichia coli during silver adaptation. Results: Fitness assays showed that as the environment changed, each mutant displayed a unique fitness profile with greatest fitness in the original selection environment. RNAseq then indicated that, in ± silver nitrate, each mutant induces a primary response that upregulates cusS, its RR cusR, and constitutively expresses the target response genes cusCFBA. This then induces a secondary response via differential expression of genes regulated by the CusR through TCRS crosstalk. Finally, each mutant undergoes fitness tuning through unique tertiary responses that result in gene expression patterns specific for the genotype, the environment and optimized for the original selection conditions. Discussion: This three-step response shows that different mutations in a single gene leads to individualized phenotypes governed by unique GxE interactions that not only contribute to transcriptional divergence but also to phenotypic plasticity.
Keywords: Escherichia coli; adaptation; fitness; gene-by-environment (GxE) interaction; two-component response systems.
Copyright © 2024 Sanders, Miller, Ahmidouch, Graves and Thomas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The Sensory Histidine Kinase CusS of Escherichia coli Senses Periplasmic Copper Ions.Microbiol Spectr. 2023 Mar 14;11(2):e0029123. doi: 10.1128/spectrum.00291-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 36916932 Free PMC article.
-
Mechanism of metal ion-induced activation of a two-component sensor kinase.Biochem J. 2019 Jan 15;476(1):115-135. doi: 10.1042/BCJ20180577. Biochem J. 2019. PMID: 30530842 Free PMC article.
-
The Structure of the Periplasmic Sensor Domain of the Histidine Kinase CusS Shows Unusual Metal Ion Coordination at the Dimeric Interface.Biochemistry. 2016 Sep 20;55(37):5296-306. doi: 10.1021/acs.biochem.6b00707. Epub 2016 Sep 12. Biochemistry. 2016. PMID: 27583660 Free PMC article.
-
Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another?Antonie Van Leeuwenhoek. 1998 Jan;73(1):35-47. doi: 10.1023/a:1000675521611. Antonie Van Leeuwenhoek. 1998. PMID: 9602277 Review.
-
The genomic determinants of genotype × environment interactions in gene expression.Trends Genet. 2013 Aug;29(8):479-87. doi: 10.1016/j.tig.2013.05.006. Epub 2013 Jun 13. Trends Genet. 2013. PMID: 23769209 Review.
Cited by
-
It Takes Two to Make a Thing Go Right: Epistasis, Two-Component Response Systems, and Bacterial Adaptation.Microorganisms. 2024 Sep 30;12(10):2000. doi: 10.3390/microorganisms12102000. Microorganisms. 2024. PMID: 39458309 Free PMC article.
References
LinkOut - more resources
Full Text Sources

