This is a preprint.
Substrate Prediction for RiPP Biosynthetic Enzymes via Masked Language Modeling and Transfer Learning
- PMID: 38463513
- PMCID: PMC10925380
Substrate Prediction for RiPP Biosynthetic Enzymes via Masked Language Modeling and Transfer Learning
Update in
-
Substrate prediction for RiPP biosynthetic enzymes via masked language modeling and transfer learning.Digit Discov. 2024 Dec 2;4(2):343-354. doi: 10.1039/d4dd00170b. eCollection 2025 Feb 12. Digit Discov. 2024. PMID: 39649639 Free PMC article.
Abstract
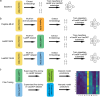
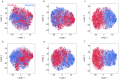
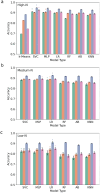
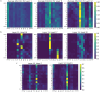
Ribosomally synthesized and post-translationally modified peptide (RiPP) biosynthetic enzymes often exhibit promiscuous substrate preferences that cannot be reduced to simple rules. Large language models are promising tools for predicting such peptide fitness landscapes. However, state-of-the-art protein language models are trained on relatively few peptide sequences. A previous study comprehensively profiled the peptide substrate preferences of LazBF (a two-component serine dehydratase) and LazDEF (a three-component azole synthetase) from the lactazole biosynthetic pathway. We demonstrated that masked language modeling of LazBF substrate preferences produced language model embeddings that improved downstream classification models of both LazBF and LazDEF substrates. Similarly, masked language modelling of LazDEF substrate preferences produced embeddings that improved the performance of classification models of both LazBF and LazDEF substrates. Our results suggest that the models learned functional forms that are transferable between distinct enzymatic transformations that act within the same biosynthetic pathway. Our transfer learning method improved performance and data efficiency in data-scarce scenarios. We then fine-tuned models on each data set and showed that the fine-tuned models provided interpretable insight that we anticipate will facilitate the design of substrate libraries that are compatible with desired RiPP biosynthetic pathways.
Figures




Similar articles
-
Substrate prediction for RiPP biosynthetic enzymes via masked language modeling and transfer learning.Digit Discov. 2024 Dec 2;4(2):343-354. doi: 10.1039/d4dd00170b. eCollection 2025 Feb 12. Digit Discov. 2024. PMID: 39649639 Free PMC article.
-
Accurate Models of Substrate Preferences of Post-Translational Modification Enzymes from a Combination of mRNA Display and Deep Learning.ACS Cent Sci. 2022 Jun 22;8(6):814-824. doi: 10.1021/acscentsci.2c00223. Epub 2022 May 26. ACS Cent Sci. 2022. PMID: 35756369 Free PMC article.
-
Promiscuous Enzymes Cooperate at the Substrate Level En Route to Lactazole A.J Am Chem Soc. 2020 Aug 12;142(32):13886-13897. doi: 10.1021/jacs.0c05541. Epub 2020 Jul 31. J Am Chem Soc. 2020. PMID: 32664727
-
Cytochromes P450 involved in bacterial RiPP biosyntheses.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuad005. doi: 10.1093/jimb/kuad005. J Ind Microbiol Biotechnol. 2023. PMID: 36931895 Free PMC article. Review.
-
Cytochromes P450 Associated with the Biosyntheses of Ribosomally Synthesized and Post-translationally Modified Peptides.ACS Bio Med Chem Au. 2023 Jul 13;3(5):371-388. doi: 10.1021/acsbiomedchemau.3c00026. eCollection 2023 Oct 18. ACS Bio Med Chem Au. 2023. PMID: 37876494 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
