Genomic and functional diversity of cultivated Bifidobacterium from human gut microbiota
- PMID: 38463766
- PMCID: PMC10923715
- DOI: 10.1016/j.heliyon.2024.e27270
Genomic and functional diversity of cultivated Bifidobacterium from human gut microbiota
Abstract
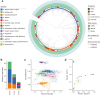
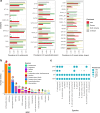
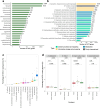
The genus Bifidobacterium widely exists in human gut and has been increasingly used as the adjuvant probiotics for the prevention and treatment of diseases. However, the functional differences of Bifidobacterium genomes from different regions of the world remain unclear. We here describe an extensive study on the genomic characteristics and function annotations of 1512 genomes (clustered to 849 non-redundant genomes) of Bifidobacterium cultured from human gut. The distribution of some carbohydrate-active enzymes varied among different Bifidobacterium species and continents. More than 36% of the genomes of B. pseudocatenulatum harbored biosynthetic gene clusters of lanthipeptide-class-iv. 99.76% of the cultivated genomes of Bifidobacterium harbored genes of bile salt hydrolase. Most genomes of B. adolescentis, and all genomes of B. dentium harbored genes involved in gamma-aminobutyric acid synthesis. B. longum subsp. infantis were characterized harboring most genes related to human milk oligosaccharide utilization. Significant differences between the distribution of antibiotic resistance genes among different species and continents revealed the importance to use antibiotics precisely in the clinical treatment. Phages infecting Bifidobacterium and horizontal gene transfers occurring in genomes of Bifidobacterium were dependent on species and region sources, and might help Bifidobacterium adapt to the environment. In addition, the distribution of Bifidobacterium in human gut was found varied from different regions of the world. This study represents a comprehensive view of characteristics and functions of genomes of cultivated Bifidobacterium from human gut, and enables clinical advances in the future.
Keywords: Bifidobacterium; Bile salt hydrolase; Gut microbiome; Horizontal gene transfers; Phages.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Exploring the Genomic Diversity and Antimicrobial Susceptibility of Bifidobacterium pseudocatenulatum in a Vietnamese Population.Microbiol Spectr. 2021 Oct 31;9(2):e0052621. doi: 10.1128/Spectrum.00526-21. Epub 2021 Sep 15. Microbiol Spectr. 2021. PMID: 34523984 Free PMC article.
-
Galacto- and Fructo-oligosaccharides Utilized for Growth by Cocultures of Bifidobacterial Species Characteristic of the Infant Gut.Appl Environ Microbiol. 2020 May 19;86(11):e00214-20. doi: 10.1128/AEM.00214-20. Print 2020 May 19. Appl Environ Microbiol. 2020. PMID: 32220841 Free PMC article.
-
Regional variation and adaptive evolution in Bifidobacterium pseudocatenulatum: Insights into genomic and functional diversity in human gut.Food Res Int. 2024 Sep;192:114840. doi: 10.1016/j.foodres.2024.114840. Epub 2024 Jul 31. Food Res Int. 2024. PMID: 39147525
-
Meta-analysis reveals different functional characteristics of human gut Bifidobacteria associated with habitual diet.Food Res Int. 2023 Aug;170:112981. doi: 10.1016/j.foodres.2023.112981. Epub 2023 May 29. Food Res Int. 2023. PMID: 37316017 Review.
-
Bosom Buddies: The Symbiotic Relationship Between Infants and Bifidobacterium longum ssp. longum and ssp. infantis. Genetic and Probiotic Features.Annu Rev Food Sci Technol. 2016;7:1-21. doi: 10.1146/annurev-food-041715-033151. Annu Rev Food Sci Technol. 2016. PMID: 26934170 Review.
Cited by
-
Gut-Brain Axis in Mood Disorders: A Narrative Review of Neurobiological Insights and Probiotic Interventions.Biomedicines. 2025 Jul 26;13(8):1831. doi: 10.3390/biomedicines13081831. Biomedicines. 2025. PMID: 40868087 Free PMC article. Review.
-
Specific microbial ratio in the gut microbiome is associated with multiple sclerosis.Proc Natl Acad Sci U S A. 2025 Mar 11;122(10):e2413953122. doi: 10.1073/pnas.2413953122. Epub 2025 Mar 3. Proc Natl Acad Sci U S A. 2025. PMID: 40030030
References
-
- Drago L., et al. Cultivable and pyrosequenced fecal microflora in centenarians and young subjects. J. Clin. Gastroenterol. 2012;46(Suppl):S81–S84. - PubMed
LinkOut - more resources
Full Text Sources

