This is a preprint.
Distinct Escherichia coli transcriptional profiles in the guts of recurrent UTI sufferers revealed by pangenome hybrid selection
- PMID: 38463963
- PMCID: PMC10925322
- DOI: 10.1101/2024.02.29.582780
Distinct Escherichia coli transcriptional profiles in the guts of recurrent UTI sufferers revealed by pangenome hybrid selection
Update in
-
Distinct Escherichia coli transcriptional profiles in the guts of recurrent UTI sufferers revealed by pangenome hybrid selection.Nat Commun. 2024 Nov 2;15(1):9466. doi: 10.1038/s41467-024-53829-7. Nat Commun. 2024. PMID: 39487120 Free PMC article.
Abstract
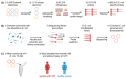
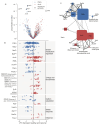
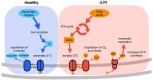
Low-abundance members of microbial communities are difficult to study in their native habitats. This includes Escherichia coli, a minor, but common inhabitant of the gastrointestinal tract and opportunistic pathogen, including of the urinary tract, where it is the primary pathogen. While multi-omic analyses have detailed critical interactions between uropathogenic Escherichia coli (UPEC) and the bladder that mediate UTI outcome, comparatively little is known about UPEC in its pre-infection reservoir, partly due to its low abundance there (<1% relative abundance). To accurately and sensitively explore the genomes and transcriptomes of diverse E. coli in gastrointestinal communities, we developed E. coli PanSelect which uses a set of probes designed to specifically recognize and capture E. coli's broad pangenome from sequencing libraries. We demonstrated the ability of E. coli PanSelect to enrich, by orders of magnitude, sequencing data from diverse E. coli using a mock community and a set of human stool samples collected as part of a cohort study investigating drivers of recurrent urinary tract infections (rUTI). Comparisons of genomes and transcriptomes between E. coli residing in the gastrointestinal tracts of women with and without a history of rUTI suggest that rUTI gut E. coli are responding to increased levels of oxygen and nitrate, suggestive of mucosal inflammation, which may have implications for recurrent disease. E. coli PanSelect is well suited for investigations of native in vivo biology of E. coli in other environments where it is at low relative abundance, and the framework described here has broad applicability to other highly diverse, low abundance organisms.
Figures


References
-
- Foxman B. (2002). Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113 Suppl 1A, 5S–13S. - PubMed
-
- Lancet The (2018). Balancing treatment with resistance in UTIs. Lancet 391, 1966. - PubMed
-
- Mediavilla J.R., Patrawalla A., Chen L., Chavda K.D., Mathema B., Vinnard C., Dever L.L., and Kreiswirth B.N. (2016). Colistin- and Carbapenem-Resistant Escherichia coli Harboring mcr-1 and blaNDM-5, Causing a Complicated Urinary Tract Infection in a Patient from the United States. MBio 7. 10.1128/mBio.01191-16. - DOI - PMC - PubMed
-
- Ajiboye R.M., Solberg O.D., Lee B.M., Raphael E., Debroy C., and Riley L.W. (2009). Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin. Infect. Dis. 49, 365–371. - PubMed
-
- Karlowsky J.A., Hoban D.J., Decorby M.R., Laing N.M., and Zhanel G.G. (2006). Fluoroquinolone-resistant urinary isolates of Escherichia coli from outpatients are frequently multidrug resistant: results from the North American Urinary Tract Infection Collaborative Alliance-Quinolone Resistance study. Antimicrob. Agents Chemother. 50, 2251–2254. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
