This is a preprint.
Functional overlap between the mammalian Sar1a and Sar1b paralogs in vivo
- PMID: 38463989
- PMCID: PMC10925261
- DOI: 10.1101/2024.02.27.582310
Functional overlap between the mammalian Sar1a and Sar1b paralogs in vivo
Update in
-
Functional overlap between the mammalian Sar1a and Sar1b paralogs in vivo.Proc Natl Acad Sci U S A. 2024 May 7;121(19):e2322164121. doi: 10.1073/pnas.2322164121. Epub 2024 Apr 30. Proc Natl Acad Sci U S A. 2024. PMID: 38687799 Free PMC article.
Abstract
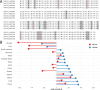
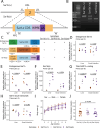
Proteins carrying a signal peptide and/or a transmembrane domain enter the intracellular secretory pathway at the endoplasmic reticulum (ER) and are transported to the Golgi apparatus via COPII vesicles or tubules. SAR1 initiates COPII coat assembly by recruiting other coat proteins to the ER membrane. Mammalian genomes encode two SAR1 paralogs, SAR1A and SAR1B. While these paralogs exhibit ~90% amino acid sequence identity, it is unknown whether they perform distinct or overlapping functions in vivo. We now report that genetic inactivation of Sar1a in mice results in lethality during mid-embryogenesis. We also confirm previous reports that complete deficiency of murine Sar1b results in perinatal lethality. In contrast, we demonstrate that deletion of Sar1b restricted to hepatocytes is compatible with survival, though resulting in hypocholesterolemia that can be rescued by adenovirus-mediated overexpression of either SAR1A or SAR1B. To further examine the in vivo function of these 2 paralogs, we genetically engineered mice with the Sar1a coding sequence replacing that of Sar1b at the endogenous Sar1b locus. Mice homozygous for this allele survive to adulthood and are phenotypically normal, demonstrating complete or near-complete overlap in function between the two SAR1 protein paralogs in mice. These data also suggest upregulation of SAR1A gene expression as a potential approach for the treatment of SAR1B deficiency (chylomicron retention disease) in humans.
Figures



Similar articles
-
Functional overlap between the mammalian Sar1a and Sar1b paralogs in vivo.Proc Natl Acad Sci U S A. 2024 May 7;121(19):e2322164121. doi: 10.1073/pnas.2322164121. Epub 2024 Apr 30. Proc Natl Acad Sci U S A. 2024. PMID: 38687799 Free PMC article.
-
Small sequence variations between two mammalian paralogs of the small GTPase SAR1 underlie functional differences in coat protein complex II assembly.J Biol Chem. 2020 Jun 19;295(25):8401-8412. doi: 10.1074/jbc.RA120.012964. Epub 2020 May 1. J Biol Chem. 2020. PMID: 32358066 Free PMC article.
-
Arabidopsis Sar1 isoforms play redundant roles in female gametophytic development.Plant Reprod. 2023 Dec;36(4):349-354. doi: 10.1007/s00497-023-00475-6. Epub 2023 Aug 3. Plant Reprod. 2023. PMID: 37535249
-
The intracellular transport of chylomicrons requires the small GTPase, Sar1b.Curr Opin Lipidol. 2004 Apr;15(2):191-7. doi: 10.1097/00041433-200404000-00012. Curr Opin Lipidol. 2004. PMID: 15017362 Review.
-
Chylomicron retention disease: genetics, biochemistry, and clinical spectrum.Curr Opin Lipidol. 2019 Apr;30(2):134-139. doi: 10.1097/MOL.0000000000000578. Curr Opin Lipidol. 2019. PMID: 30640893 Review.
References
-
- Zanetti G., Pahuja K. B., Studer S., Shim S., Schekman R., COPII and the regulation of protein sorting in mammals. Nat Cell Biol 14, 20–28 (2011). - PubMed
-
- Barlowe C., Helenius A., Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu Rev Cell Dev Biol 32, 197–222 (2016). - PubMed
-
- Novick P., Field C., Schekman R., Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215 (1980). - PubMed
-
- Barlowe C. et al., COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895--907 (1994). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
