This is a preprint.
Suppressing APOE4-induced mortality and cellular damage by targeting VHL
- PMID: 38464138
- PMCID: PMC10925324
- DOI: 10.1101/2024.02.28.582664
Suppressing APOE4-induced mortality and cellular damage by targeting VHL
Abstract
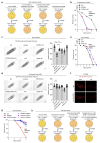
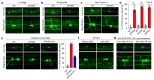
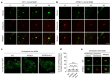
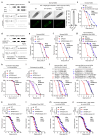
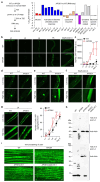
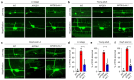
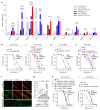
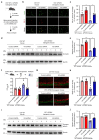
Mortality rate increases with age and can accelerate upon extrinsic or intrinsic damage to individuals. Identifying factors and mechanisms that curb population mortality rate has wide-ranging implications. Here, we show that targeting the VHL-1 (Von Hippel-Lindau) protein suppresses C. elegans mortality caused by distinct factors, including elevated reactive oxygen species, temperature, and APOE4, the genetic variant that confers high risks of neurodegeneration in Alzheimer's diseases and all-cause mortality in humans. These mortality factors are of different physical-chemical nature, yet result in similar cellular dysfunction and damage that are suppressed by deleting VHL-1. Stabilized HIF-1 (hypoxia inducible factor), a transcription factor normally targeted for degradation by VHL-1, recapitulates the protective effects of deleting VHL-1. HIF-1 orchestrates a genetic program that defends against mitochondrial abnormalities, excess oxidative stress, cellular proteostasis dysregulation, and endo-lysosomal rupture, key events that lead to mortality. Genetic Vhl inhibition also alleviates cerebral vascular injury and synaptic lesions in APOE4 mice, supporting an evolutionarily conserved mechanism. Collectively, we identify the VHL-HIF axis as a potent modifier of APOE4 and mortality and propose that targeting VHL-HIF in non-proliferative animal tissues may suppress tissue injuries and mortality by broadly curbing cellular damage.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures






Similar articles
-
Suppressing APOE4-induced neural pathologies by targeting the VHL-HIF axis.Proc Natl Acad Sci U S A. 2025 Feb 4;122(5):e2417515122. doi: 10.1073/pnas.2417515122. Epub 2025 Jan 28. Proc Natl Acad Sci U S A. 2025. PMID: 39874294 Free PMC article.
-
Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease.Cancer Res. 2002 Jul 1;62(13):3803-11. Cancer Res. 2002. PMID: 12097293
-
Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function.Oncogene. 2000 Nov 16;19(48):5435-43. doi: 10.1038/sj.onc.1203938. Oncogene. 2000. PMID: 11114720
-
Molecular targets from VHL studies into the oxygen-sensing pathway.Curr Cancer Drug Targets. 2005 Aug;5(5):345-56. doi: 10.2174/1568009054629672. Curr Cancer Drug Targets. 2005. PMID: 16101382 Review.
-
The von Hippel-Lindau tumor suppressor gene and kidney cancer.Clin Cancer Res. 2004 Sep 15;10(18 Pt 2):6290S-5S. doi: 10.1158/1078-0432.CCR-sup-040025. Clin Cancer Res. 2004. PMID: 15448019 Review.
References
-
- Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., and Roses A.D. (1993). Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A 90, 1977–1981. 10.1073/pnas.90.5.1977. - DOI - PMC - PubMed
-
- Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., and Pericak-Vance M.A. (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. 10.1126/science.8346443. - DOI - PubMed
-
- Bejanin A., lulita M.F., Vilaplana E., Carmona-Iragui M., Benejam B., Videla L., Barroeta I., Fernandez S., Altuna M., Pegueroles J., et al. (2021). Association of Apolipoprotein E ε4 Allele With Clinical and Multimodal Biomarker Changes of Alzheimer Disease in Adults With Down Syndrome. JAMA Neurol 78, 937–947. 10.1001/jamaneurol.2021.1893. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
