This is a preprint.
Targeting Iron - Respiratory Reciprocity Promotes Bacterial Death
- PMID: 38464199
- PMCID: PMC10925246
- DOI: 10.1101/2024.03.01.582947
Targeting Iron - Respiratory Reciprocity Promotes Bacterial Death
Update in
-
Lacritin cleavage-potentiated targeting of iron - respiratory reciprocity promotes bacterial death.J Biol Chem. 2025 May;301(5):108455. doi: 10.1016/j.jbc.2025.108455. Epub 2025 Mar 26. J Biol Chem. 2025. PMID: 40154612 Free PMC article.
Abstract
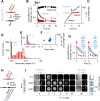
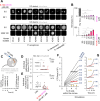
Discovering new bacterial signaling pathways offers unique antibiotic strategies. Here, through an unbiased resistance screen of 3,884 gene knockout strains, we uncovered a previously unknown non-lytic bactericidal mechanism that sequentially couples three transporters and downstream transcription to lethally suppress respiration of the highly virulent P. aeruginosa strain PA14 - one of three species on the WHO's 'Priority 1: Critical' list. By targeting outer membrane YaiW, cationic lacritin peptide 'N-104' translocates into the periplasm where it ligates outer loops 4 and 2 of the inner membrane transporters FeoB and PotH, respectively, to suppress both ferrous iron and polyamine uptake. This broadly shuts down transcription of many biofilm-associated genes, including ferrous iron-dependent TauD and ExbB1. The mechanism is innate to the surface of the eye and is enhanced by synergistic coupling with thrombin peptide GKY20. This is the first example of an inhibitor of multiple bacterial transporters.
Conflict of interest statement
DECLARATION OF INTERESTS GWL is cofounder and CSO of TearSolutions, Inc; and cofounder and CTO of IsletRegen, LLC. Other authors declare no competing interests.
Figures





Similar articles
-
Lacritin cleavage-potentiated targeting of iron - respiratory reciprocity promotes bacterial death.J Biol Chem. 2025 May;301(5):108455. doi: 10.1016/j.jbc.2025.108455. Epub 2025 Mar 26. J Biol Chem. 2025. PMID: 40154612 Free PMC article.
-
PQS and pyochelin in Pseudomonas aeruginosa share inner membrane transporters to mediate iron uptake.Microbiol Spectr. 2024 Feb 6;12(2):e0325623. doi: 10.1128/spectrum.03256-23. Epub 2024 Jan 3. Microbiol Spectr. 2024. PMID: 38171001 Free PMC article.
-
Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter.Microbiology (Reading). 2009 Jan;155(Pt 1):305-315. doi: 10.1099/mic.0.023531-0. Microbiology (Reading). 2009. PMID: 19118371
-
Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways.Amino Acids. 2013 May;44(5):1267-77. doi: 10.1007/s00726-013-1468-2. Epub 2013 Feb 27. Amino Acids. 2013. PMID: 23443998 Review.
-
Toward a mechanistic understanding of Feo-mediated ferrous iron uptake.Metallomics. 2018 Jul 18;10(7):887-898. doi: 10.1039/c8mt00097b. Metallomics. 2018. PMID: 29953152 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
