This is a preprint.
Meta-analysis of the human gut microbiome uncovers shared and distinct microbial signatures between diseases
- PMID: 38464323
- PMCID: PMC10925178
- DOI: 10.1101/2024.02.27.582333
Meta-analysis of the human gut microbiome uncovers shared and distinct microbial signatures between diseases
Update in
-
Meta-analysis of the human gut microbiome uncovers shared and distinct microbial signatures between diseases.mSystems. 2024 Aug 20;9(8):e0029524. doi: 10.1128/msystems.00295-24. Epub 2024 Jul 30. mSystems. 2024. PMID: 39078158 Free PMC article.
Abstract
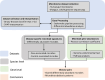
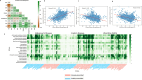
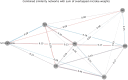
Microbiome studies have revealed gut microbiota's potential impact on complex diseases. However, many studies often focus on one disease per cohort. We developed a meta-analysis workflow for gut microbiome profiles and analyzed shotgun metagenomic data covering 11 diseases. Using interpretable machine learning and differential abundance analysis, our findings reinforce the generalization of binary classifiers for Crohn's disease (CD) and colorectal cancer (CRC) to hold-out cohorts and highlight the key microbes driving these classifications. We identified high microbial similarity in disease pairs like CD vs ulcerative colitis (UC), CD vs CRC, Parkinson's disease vs type 2 diabetes (T2D), and schizophrenia vs T2D. We also found strong inverse correlations in Alzheimer's disease vs CD and UC. These findings detected by our pipeline provide valuable insights into these diseases.
Keywords: complex human diseases; disease similarity; meta-analysis; microbiome.
Conflict of interest statement
DISCLOSURE STATEMENT The authors declare that there is no conflict of interest.
Figures


References
-
- Allaband C, McDonald D, Vázquez-Baeza Y, Minich JJ, Tripathi A, Brenner DA, Loomba R, Smarr L, Sandborn WJ, Schnabl B, Dorrestein P, Zarrinpar A, Knight R. 2019. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin Gastroenterol Hepatol 17:218–230. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
