This is a preprint.
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19
- PMID: 38464327
- PMCID: PMC10925244
- DOI: 10.1101/2024.02.27.582110
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19
Update in
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.Microbiol Spectr. 2025 May 6;13(5):e0182924. doi: 10.1128/spectrum.01829-24. Epub 2025 Mar 25. Microbiol Spectr. 2025. PMID: 40130852 Free PMC article.
Abstract
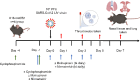
Objectives: Immunocompromised individuals are susceptible to severe COVID-19 and potentially contribute to the emergence of variants with altered pathogenicity due to persistent infection. This study investigated the impact of immunosuppression on SARS-CoV-2 infection in k18-hACE2 mice and the effectiveness of antiviral treatments in this context during the first 7 days of infection.
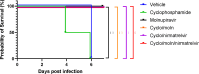
Methods: Mice were immunosuppressed using cyclophosphamide and infected with a B daughter lineage of SARS-CoV-2. Molnupiravir and nirmatrelvir, alone and in combination, were administered and viral load and viral sequence diversity was assessed.
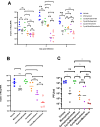
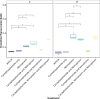
Results: Treatment of infected but immune compromised mice with both compounds either singly or in combination resulted in decreased viral loads and pathological changes compared to untreated animals. Treatment also abrogated infection of neuronal tissue. However, no consistent changes in the viral consensus sequence were observed, except for the emergence of the S:H655Y mutation. Molnupiravir, but not nirmatrelvir or immunosuppression alone, increased the transition/transversion (Ts/Tv) ratio, representative of G>A and C>U mutations and this increase was not altered by the co-administration of nirmatrelvir with molnupiravir.Notably, immunosuppression itself did not appear to promote the emergence of mutational characteristic of variants of concern (VOCs).
Conclusions: Further investigations are warranted to fully understand the role of immunocompromised individuals in VOC development, especially by taking persistence into consideration, and to inform optimised public health strategies. It is more likely that immunodeficiency promotes viral persistence but does not necessarily lead to substantial consensus-level changes in the absence of antiviral selection pressure. Consistent with mechanisms of action, molnupiravir showed a stronger mutagenic effect than nirmatrelvir in this model.
Keywords: COVID-19; Molnupiravir; Nirmatrelvir; Paxlovid; SARS-CoV-2; immunocompromised; intra-host evolution.
Conflict of interest statement
Transparency Declaration A.O. is a director of Tandem Nano Ltd and co-inventor of patents relating to drug delivery. A.O. has been co-investigator on funding received by the University of Liverpool from ViiV Healthcare and Gilead Sciences in the past 3 years unrelated to COVID-19. A.O. has received personal fees from Gilead and Assembly Biosciences in the past 3 years, also unrelated to COVID-19. JPS has received funding from ENA respiratory Pty Ltd, Bicycle Tx Ltd, and Infex Therapeutics Ltd unrelated to this study. R.P.R. is an employee at TopMD Precision Medicine Ltd. No other conflicts are declared by the authors.
Figures




Similar articles
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.Microbiol Spectr. 2025 May 6;13(5):e0182924. doi: 10.1128/spectrum.01829-24. Epub 2025 Mar 25. Microbiol Spectr. 2025. PMID: 40130852 Free PMC article.
-
Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice.Antiviral Res. 2022 Dec;208:105430. doi: 10.1016/j.antiviral.2022.105430. Epub 2022 Oct 6. Antiviral Res. 2022. PMID: 36209984 Free PMC article.
-
Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study.Lancet Microbe. 2024 May;5(5):e452-e458. doi: 10.1016/S2666-5247(23)00393-2. Epub 2024 Mar 22. Lancet Microbe. 2024. PMID: 38527471
-
Viral target and metabolism-based rationale for combined use of recently authorized small molecule COVID-19 medicines: Molnupiravir, nirmatrelvir, and remdesivir.Fundam Clin Pharmacol. 2023 Aug;37(4):726-738. doi: 10.1111/fcp.12889. Epub 2023 Mar 25. Fundam Clin Pharmacol. 2023. PMID: 36931725 Free PMC article. Review.
-
Molnupiravir and Nirmatrelvir-Ritonavir: Oral Coronavirus Disease 2019 Antiviral Drugs.Clin Infect Dis. 2023 Jan 6;76(1):165-171. doi: 10.1093/cid/ciac180. Clin Infect Dis. 2023. PMID: 35245942 Free PMC article. Review.
References
-
- Moore S. C., Penrice-Randal R., Alruwaili M., Randle N., Armstrong S., Hartley C., Haldenby S., Dong X., Alrezaihi A., Almsaud M., Bentley E., Clark J., Garcia-Dorival I., Gilmore P., Han X., Jones B., Luu L., Sharma P., Shawli G., Sun Y., Zhao Q., Pullan S. T., Carter D. P., Bewley K., Dunning J., Zhou E. M., Solomon T., Beadsworth M., Cruise J., Crook D. W., Matthews D. A., Davidson A. D., Mahmood Z., Aljabr W., Druce J., Vipond R., Ng L., Renia L., Openshaw P. J. M., Baillie J. K., Carroll M. W., Stewart J., Darby A., Semple M., Turtle L. & Hiscox J. A. Amplicon-Based Detection and Sequencing of SARS-CoV-2 in Nasopharyngeal Swabs from Patients With COVID-19 and Identification of Deletions in the Viral Genome That Encode Proteins Involved in Interferon Antagonism. Viruses 12 (2020). 10.3390/v12101164 - DOI - PMC - PubMed
-
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E. E., Bhattacharya T., Foley B., Hastie K. M., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., Sheffield C.-G. G., McDanal C., Perez L. G., Tang H., Moon-Walker A., Whelan S. P., LaBranche C. C., Saphire E. O. & Montefiori D. C. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827 e819 (2020). 10.1016/j.cell.2020.06.043 - DOI - PMC - PubMed
-
- Goldswain H., Dong X., Penrice-Randal R., Alruwaili M., Shawli G. T., Prince T., Williamson M. K., Raghwani J., Randle N., Jones B., Donovan-Banfield I. a., Salguero F. J., Tree J. A., Hall Y., Hartley C., Erdmann M., Bazire J., Jearanaiwitayakul T., Semple M. G., Openshaw P. J. M., Baillie J. K., Baillie J. K., Semple M. G., Openshaw P. J. M., Carson G., Alex B., Andrikopoulos P., Bach B., Barclay W. S., Bogaert D., Chand M., Chechi K., Cooke G. S., da Silva Filipe A., de Silva T., Docherty A. B., dos Santos Correia G., Dumas M.-E., Dunning J., Fletcher T., Green C. A., Greenhalf W., Griffin J. L., Gupta R. K., Harrison E. M., Hiscox J. A., Ho A. Y. W., Horby P. W., Ijaz S., Khoo S., Klenerman P., Law A., Lewis M. R., Liggi S., Lim W. S., Maslen L., Mentzer A. J., Merson L., Meynert A. M., Moore S. C., Noursadeghi M., Olanipekun M., Osagie A., Palmarini M., Palmieri C., Paxton W. A., Pollakis G., Price N., Rambaut A., Robertson D. L., Russell C. D., SanchoShimizu V., Sands C. J., Scott J. T., Sigfrid L., Solomon T., Sriskandan S., Stuart D., Summers C., Swann O. V., Takats Z., Takis P., Tedder R. S., Thompson A. A. R., Thomson E. C., Thwaites R. S., Turtle L. C. W., Zambon M., Hardwick H., Donohue C., Griffiths F., Oosthuyzen W., Donegan C., Spencer R. G., Norman L., Pius R., Drake T. M., Fairfield C. J., Knight S. R., McLean K. A., Murphy D., Shaw C. A., Dalton J., Girvan M., Saviciute E., Roberts S., Harrison J., Marsh L., Connor M., Halpin S., Jackson C., Gamble C., Plotkin D., Lee J., Leeming G., Law A., Wham M., Clohisey S., Hendry R., Scott-Brown J., Shaw V., McDonald S. E., Keating S., Ahmed K. A., Armstrong J. A., Ashworth M., Asiimwe I. G., Bakshi S., Barlow S. L., Booth L., Brennan B., Bullock K., Catterall B. W. A., Clark J. J., Clarke E. A., Cole S., Cooper L., Cox H., Davis C., Dincarslan O., Dunn C., Dyer P., Elliott A., Evans A., Finch L., Fisher L. W. S., Foster T., Garcia-Dorival I., Gunning P., Jensen R. L., Jones C. B., Jones T. R., Khandaker S., King K., Kiy R. T., Koukorava C., Lake A., Lant S., Latawiec D., Lavelle-Langham L., Lefteri D., Lett L., Livoti L. A., Mancini M., McDonald S., McEvoy L., McLauchlan J., Metelmann S., Miah N. S., Middleton J., Mitchell J., Moore S. C., Murphy E. G., Pilgrim J., Reynolds W., Ridley P. M., Sales D., Shaw V. E., Shears R. K., Small B., Subramaniam K. S., Szemiel A., Taggart A., Tanianis-Hughes J., Thomas J., Trochu E., van Tonder L., Wilcock E., Zhang J. E., Flaherty L., Maziere N., Cass E., Carracedo A. D., Carlucci N., Holmes A., Massey H., Murphy L., McCafferty S., Clark R., Fawkes A., Morrice K., Maclean A., Wrobel N., Donnelly L., Coutts A., Hafezi K., MacGillivray L., Gilchrist T., Adeniji K., Agranoff D., Agwuh K., Ail D., Aldera E. L., Alegria A., Allen S., Angus B., Ashish A., Atkinson D., Bari S., Barlow G., Barnass S., Barrett N., Bassford C., Basude S., Baxter D., Beadsworth M., Bernatoniene J., Berridge J., Berry C., Best N., Bothma P., Chadwick D., Brittain-Long R., Bulteel N., Burden T., Burtenshaw A., Caruth V., Chadwick D., Chambler D., Chee N., Child J., Chukkambotla S., Clark T., Collini P., Cosgrove C., Cupitt J., Cutino-Moguel M.-T., Dark P., Dawson C., Dervisevic S., Donnison P., Douthwaite S., Drummond A., DuRand I., Dushianthan A., Dyer T., Evans C., Eziefula C., Fegan C., Finn A., Fullerton D., Garg S., Garg S., Garg A., Gkrania-Klotsas E., Godden J., Goldsmith A., Graham C., Hardy E., Hartshorn S., Harvey D., Havalda P., Hawcutt D. B., Hobrok M., Hodgson L., Hormis A., Jacobs M., Jain S., Jennings P., Kaliappan A., Kasipandian V., Kegg S., Kelsey M., Kendall J., Kerrison C., Kerslake I., Koch O., Koduri G., Koshy G., Laha S., Laird S., Larkin S., Leiner T., Lillie P., Limb J., Linnett V., Little J., Lyttle M., MacMahon M. & Investigators I. C. The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection. Genome Biology 24, 47 (2023). 10.1186/s13059-023-02881-5 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
