Selecting Monoclonal Cell Lineages from Somatic Reprogramming Using Robotic-Based Spatial-Restricting Structured Flow
- PMID: 38464498
- PMCID: PMC10923610
- DOI: 10.34133/research.0338
Selecting Monoclonal Cell Lineages from Somatic Reprogramming Using Robotic-Based Spatial-Restricting Structured Flow
Abstract
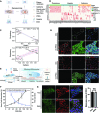
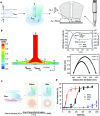
Somatic cell reprogramming generates induced pluripotent stem cells (iPSCs), which serve as a crucial source of seed cells for personalized disease modeling and treatment in regenerative medicine. However, the process of reprogramming often causes substantial lineage manipulations, thereby increasing cellular heterogeneity. As a consequence, the process of harvesting monoclonal iPSCs is labor-intensive and leads to decreased reproducibility. Here, we report the first in-house developed robotic platform that uses a pin-tip-based micro-structure to manipulate radial shear flow for automated monoclonal iPSC colony selection (~1 s) in a non-invasive and label-free manner, which includes tasks for somatic cell reprogramming culturing, medium changes; time-lapse-based high-content imaging; and iPSCs monoclonal colony detection, selection, and expansion. Throughput-wise, this automated robotic system can perform approximately 24 somatic cell reprogramming tasks within 50 days in parallel via a scheduling program. Moreover, thanks to a dual flow-based iPSC selection process, the purity of iPSCs was enhanced, while simultaneously eliminating the need for single-cell subcloning. These iPSCs generated via the dual processing robotic approach demonstrated a purity 3.7 times greater than that of the conventional manual methods. In addition, the automatically produced human iPSCs exhibited typical pluripotent transcriptional profiles, differentiation potential, and karyotypes. In conclusion, this robotic method could offer a promising solution for the automated isolation or purification of lineage-specific cells derived from iPSCs, thereby accelerating the development of personalized medicines.
Copyright © 2024 Xueping Chen et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



Similar articles
-
Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells.Nat Methods. 2015 Sep;12(9):885-92. doi: 10.1038/nmeth.3507. Epub 2015 Aug 3. Nat Methods. 2015. PMID: 26237226
-
Optimized and Scalable Precoating-Free Reprogramming of Human Peripheral Blood Mononuclear Cells into iPSCs.Curr Protoc. 2024 Jan;4(1):e979. doi: 10.1002/cpz1.979. Curr Protoc. 2024. PMID: 38265186
-
An Overview on Promising Somatic Cell Sources Utilized for the Efficient Generation of Induced Pluripotent Stem Cells.Stem Cell Rev Rep. 2021 Dec;17(6):1954-1974. doi: 10.1007/s12015-021-10200-3. Epub 2021 Jun 7. Stem Cell Rev Rep. 2021. PMID: 34100193 Review.
-
Reverse engineering human neurodegenerative disease using pluripotent stem cell technology.Brain Res. 2016 May 1;1638(Pt A):30-41. doi: 10.1016/j.brainres.2015.09.023. Epub 2015 Sep 28. Brain Res. 2016. PMID: 26423934 Free PMC article. Review.
-
Development of a Modular Automated System for Maintenance and Differentiation of Adherent Human Pluripotent Stem Cells.SLAS Discov. 2017 Sep;22(8):1016-1025. doi: 10.1177/2472555217696797. Epub 2017 Mar 13. SLAS Discov. 2017. PMID: 28287872
Cited by
-
Mammalian Ste20-like kinase 1 regulates AMPK to mitigate the progression of non-alcoholic fatty liver disease.Eur J Med Res. 2025 Apr 17;30(1):296. doi: 10.1186/s40001-025-02557-9. Eur J Med Res. 2025. PMID: 40247356 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

