Brucella abortus triggers the differential expression of immunomodulatory lncRNAs in infected murine macrophages
- PMID: 38464511
- PMCID: PMC10921354
- DOI: 10.3389/fimmu.2024.1352306
Brucella abortus triggers the differential expression of immunomodulatory lncRNAs in infected murine macrophages
Abstract
Introduction: The lncRNAs (long non-coding RNAs) are the most diverse group of non-coding RNAs and are involved in most biological processes including the immune response. While some of them have been recognized for their influence on the regulation of inflammatory activity, little is known in the context of infection by Brucella abortus, a pathogen that presents significant challenges due to its ability to manipulate and evade the host immune system. This study focuses on characterize the expression profile of LincRNA-cox2, Lethe, lincRNA-EPS, Malat1 and Gas5 during infection of macrophages by B. abortus.
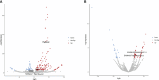
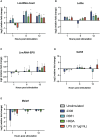
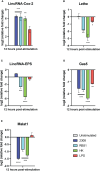
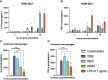
Methods: Using public raw RNA-seq datasets we constructed for a lncRNA expression profile in macrophages Brucella-infected. In addition, from public RNA-seq raw datasets of RAW264.7 cells infected with B. abortus we constructed a transcriptomic profile of lncRNAs in order to know the expression of the five immunomodulating lncRNAs studied here at 8 and 24 h post-infection. Finally, we performed in vitro infection assays in RAW264.7 cells and peritoneal macrophages to detect by qPCR changes in the expression of these lncRNAs at first 12 hours post infection, a key stage in the infection cycle where Brucella modulates the immune response to survive.
Results: Our results demonstrate that infection of macrophages with Brucella abortus, induces significant changes in the expression of LincRNA-Cox2, Lethe, LincRNA-EPS, Gas5, and Malat1.
Discussion: The change in the expression profile of these immunomodulatory lncRNAs in response to infection, suggest a potential involvement in the immune evasion strategy employed by Brucella to facilitate its intracellular survival.
Keywords: brucella abortus; gene expression; immunomodulation; lncRNA; macrophages infection.
Copyright © 2024 Flores-Concha, Gómez, Soto-Shara, Molina, Coloma-Rivero, Montero, Ferrari and Oñate.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

