Potential immune evasion of the severe acute respiratory syndrome coronavirus 2 Omicron variants
- PMID: 38464527
- PMCID: PMC10924305
- DOI: 10.3389/fimmu.2024.1339660
Potential immune evasion of the severe acute respiratory syndrome coronavirus 2 Omicron variants
Abstract
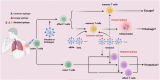
Coronavirus disease 2019 (COVID-19), which is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a global pandemic. The Omicron variant (B.1.1.529) was first discovered in November 2021 in specimens collected from Botswana, South Africa. Omicron has become the dominant variant worldwide, and several sublineages or subvariants have been identified recently. Compared to those of other mutants, the Omicron variant has the most highly expressed amino acid mutations, with almost 60 mutations throughout the genome, most of which are in the spike (S) protein, especially in the receptor-binding domain (RBD). These mutations increase the binding affinity of Omicron variants for the ACE2 receptor, and Omicron variants may also lead to immune escape. Despite causing milder symptoms, epidemiological evidence suggests that Omicron variants have exceptionally higher transmissibility, higher rates of reinfection and greater spread than the prototype strain as well as other preceding variants. Additionally, overwhelming amounts of data suggest that the levels of specific neutralization antibodies against Omicron variants decrease in most vaccinated populations, although CD4+ and CD8+ T-cell responses are maintained. Therefore, the mechanisms underlying Omicron variant evasion are still unclear. In this review, we surveyed the current epidemic status and potential immune escape mechanisms of Omicron variants. Especially, we focused on the potential roles of viral epitope mutations, antigenic drift, hybrid immunity, and "original antigenic sin" in mediating immune evasion. These insights might supply more valuable concise information for us to understand the spreading of Omicron variants.
Keywords: Omicron variant; SARS-CoV-2; hybrid immunity; immune evasion; original antigenic sin.
Copyright © 2024 Chen, He, Liu, Shang and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages.Signal Transduct Target Ther. 2022 Jul 19;7(1):241. doi: 10.1038/s41392-022-01105-9. Signal Transduct Target Ther. 2022. PMID: 35853878 Free PMC article. Review.
-
Evolution of Immune Evasion and Host Range Expansion by the SARS-CoV-2 B.1.1.529 (Omicron) Variant.mBio. 2023 Apr 25;14(2):e0041623. doi: 10.1128/mbio.00416-23. Epub 2023 Apr 3. mBio. 2023. PMID: 37010428 Free PMC article.
-
Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies.Front Immunol. 2022 Jan 24;12:830527. doi: 10.3389/fimmu.2021.830527. eCollection 2021. Front Immunol. 2022. PMID: 35140714 Free PMC article.
-
Omicron variant (B.1.1.529) and its sublineages: What do we know so far amid the emergence of recombinant variants of SARS-CoV-2?Biomed Pharmacother. 2022 Oct;154:113522. doi: 10.1016/j.biopha.2022.113522. Epub 2022 Aug 15. Biomed Pharmacother. 2022. PMID: 36030585 Free PMC article. Review.
Cited by
-
Genomic Epidemiology of the Main SARS-CoV-2 Variants Circulating in Italy During the Omicron Era.J Med Virol. 2025 Feb;97(2):e70215. doi: 10.1002/jmv.70215. J Med Virol. 2025. PMID: 39936851 Free PMC article.
-
The Inhibiting Effect of GB-2, (+)-Catechin, Theaflavin, and Theaflavin 3-Gallate on Interaction between ACE2 and SARS-CoV-2 EG.5.1 and HV.1 Variants.Int J Mol Sci. 2024 Aug 31;25(17):9498. doi: 10.3390/ijms25179498. Int J Mol Sci. 2024. PMID: 39273444 Free PMC article.
-
Fusion protein-based COVID-19 vaccines exemplified by a chimeric vaccine based on a single fusion protein (W-PreS-O).Front Immunol. 2025 Jan 28;16:1452814. doi: 10.3389/fimmu.2025.1452814. eCollection 2025. Front Immunol. 2025. PMID: 39935478 Free PMC article.
-
Identifying the effectiveness of face mask in a large population with a network-based fluid model.PLoS One. 2025 Jun 10;20(6):e0324229. doi: 10.1371/journal.pone.0324229. eCollection 2025. PLoS One. 2025. PMID: 40493681 Free PMC article.
-
4D-DIA Proteomics Uncovers New Insights into Host Salivary Response Following SARS-CoV-2 Omicron Infection.J Proteome Res. 2025 Feb 7;24(2):499-514. doi: 10.1021/acs.jproteome.4c00630. Epub 2025 Jan 13. J Proteome Res. 2025. PMID: 39803891 Free PMC article.
References
-
- dashboard . WcC. (2024). Available at: https://covid19.who.int/.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

