Establishment and application of a rapid molecular diagnostic platform for the isothermal visual amplification of group B Streptococcus based on recombinase polymerase
- PMID: 38465235
- PMCID: PMC10920233
- DOI: 10.3389/fcimb.2024.1281827
Establishment and application of a rapid molecular diagnostic platform for the isothermal visual amplification of group B Streptococcus based on recombinase polymerase
Abstract
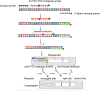
With growing concerns about Group B streptococcal (GBS) infections and their adverse effects on perinatal pregnancies, including infection, premature delivery, neonatal septicemia, and meningitis, it is urgent to promote GBS screening at all pregnancy stages. The purpose of this study is to establish a device-independent, fast, sensitive, and visual GBS detection method. Taking advantage of the characteristics of the recombinase polymerase isothermal amplification (RPA), the activity of the nfo nuclease cleavage base analog (tetrahydrofuran, THF) site, and the advantages of visual reading of the lateral flow chromatography strip (LFS), a GBS detection method was developed. This method focused on the conservative region of the Christie-Atkins-Munch-Petersen factor encoded by the cfb gene, a virulence gene specific to GBS. Two forward primers, two biotin-labeled reverse primers, and one fluorescein isothiocyanate (FITC)-labeled and C3spacer-blocked probe were designed. The study involved optimizing the primer pair and probe combination, determining the optimal reaction temperature and time, evaluating specificity, analyzing detection limits, and testing the method on 87 vaginal swabs from perinatal pregnant women. The results showed that the visual detection method of GBS-RPA-LFS, using the cfb-F1/R2/P1 primer probe, could detect GBS within 15 min at the temperature ranging from 39°C to 42°C. Furthermore, the method specifically amplified only GBS, without cross-reacting with pathogens like Lactobacillus iners, Lactobacillus crispatus, Candida albicans, Listeria monocytogenes, Yersinia enterocolitica, Klebsiella Pneumoniae, Enterobacter cloacae, Citrobacter freundii, Vibrio alginolyticus, Vibrio parahaemolyticus, Salmonella typhimurium, Staphylococcus aureus, Pseudomonas aeruginosa, or Trichomonas vaginalis. It could detect a minimum of 100 copies per reaction. In clinical 98 samples of vaginal swabs from pregnant women, the agreement rate between the GBS-RPA-LFS method and TaqMan real-time fluorescence quantification method was 95.92%. In conclusion, this study successfully established a combined RPA and LFS GBS in situ detection platform, with short reaction time, high sensitivity, high specificity, portability, and device independence, providing a feasible strategy for clinical GBS screening.
Keywords: cfb; group B streptococcus; isothermal amplification; molecular diagnostic methods; recombinase polymerase.
Copyright © 2024 Liu, Wang, Chu, Min, Sun, Zhang and Meng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Establishment and application of a point-of-care testing and diagnosis method for early immediate expression gene IE1 of cytomegalovirus in maternal urine based on isothermal amplification.Virus Res. 2023 Nov;337:199229. doi: 10.1016/j.virusres.2023.199229. Epub 2023 Oct 7. Virus Res. 2023. PMID: 37769815 Free PMC article.
-
Establishment and application of a rapid visual diagnostic method for Streptococcus agalactiae based on recombinase polymerase amplification and lateral flow strips.Sci Rep. 2024 May 2;14(1):10064. doi: 10.1038/s41598-024-56138-7. Sci Rep. 2024. PMID: 38698011 Free PMC article.
-
Establishment of a rapid diagnosis method for Candida glabrata based on the ITS2 gene using recombinase polymerase amplification combined with lateral flow strips.Front Cell Infect Microbiol. 2022 Jul 28;12:953302. doi: 10.3389/fcimb.2022.953302. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35967865 Free PMC article.
-
Rapid Detection of Klebsiella pneumoniae Carrying Virulence Gene rmpA2 by Recombinase Polymerase Amplification Combined With Lateral Flow Strips.Front Cell Infect Microbiol. 2022 May 19;12:877649. doi: 10.3389/fcimb.2022.877649. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35663473 Free PMC article.
-
Rapid and reliable loop-mediated isothermal amplification method for detecting Streptococcus agalactiae.Jpn J Infect Dis. 2013;66(6):546-8. doi: 10.7883/yoken.66.546. Jpn J Infect Dis. 2013. PMID: 24270149
Cited by
-
Evaluation of a novel liquid-solid biphasic differential medium for screening of group B streptococcus in perinatal women.Microbiol Spectr. 2025 Jun 3;13(6):e0014025. doi: 10.1128/spectrum.00140-25. Epub 2025 May 13. Microbiol Spectr. 2025. PMID: 40358227 Free PMC article.
-
CAMP-negative Streptococcus agalactiae strains exhibited complete or partial chromosomal deletions of the CAMP-factor encoding gene cfb.Microbiol Spectr. 2025 Apr 9;13(5):e0325724. doi: 10.1128/spectrum.03257-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40202309 Free PMC article.
References
-
- Abd El Wahed A., El-Deeb A., El-Tholoth M., Abd El Kader H., Ahmed A., Hassan S., et al. . (2013). A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PloS One 8 (8), e71642. doi: 10.1371/journal.pone.0071642 - DOI - PMC - PubMed
-
- Awwad E., Srour M., Hasan S., Khatib S. (2022). Molecular determination, serotyping, antibiotic profile and virulence factors of group B Streptococcus isolated from invasive patients at Arabcare Hospital Laboratory, Palestine. Am. J. Infect. Control 50 (8), 934–940. doi: 10.1016/j.ajic.2021.12.006 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

