Tracing histoplasmosis genomic epidemiology and species occurrence across the USA
- PMID: 38465644
- PMCID: PMC10930103
- DOI: 10.1080/22221751.2024.2315960
Tracing histoplasmosis genomic epidemiology and species occurrence across the USA
Abstract
ABSTRACTHistoplasmosis is an endemic mycosis in North America frequently reported along the Ohio and Mississippi River Valleys, although autochthonous cases occur in non-endemic areas. In the United States, the disease is provoked by two genetically distinct clades of Histoplasma capsulatum sensu lato, Histoplasma mississippiense (Nam1) and H. ohiense (Nam2). To bridge the molecular epidemiological gap, we genotyped 93 Histoplasma isolates (62 novel genomes) including clinical, environmental, and veterinarian samples from a broader geographical range by whole-genome sequencing, followed by evolutionary and species niche modelling analyses. We show that histoplasmosis is caused by two major lineages, H. ohiense and H. mississippiense; with sporadic cases caused by H. suramericanum in California and Texas. While H. ohiense is prevalent in eastern states, H. mississipiense was found to be prevalent in the central and western portions of the United States, but also geographically overlapping in some areas suggesting that these species might co-occur. Species Niche Modelling revealed that H. ohiense thrives in places with warmer and drier conditions, while H. mississippiense is endemic to areas with cooler temperatures and more precipitation. In addition, we predicted multiple areas of secondary contact zones where the two species co-occur, potentially facilitating gene exchange and hybridization. This study provides the most comprehensive understanding of the genomic epidemiology of histoplasmosis in the USA and lays a blueprint for the study of invasive fungal diseases.
Keywords: Histoplasma mississippiense; Histoplasma ohiense; Histoplasmosis; genomics; molecular epidemiology; species distribution modelling.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

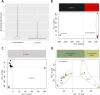

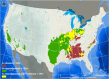
References
-
- Linder KA, Kauffman CA.. Histoplasmosis: epidemiology, diagnosis, and clinical manifestations. Curr Fungal Infect Rep. 2019/09/01;13(3):120–128. doi: 10.1007/s12281-019-00341-x - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
