Landscape of IGH germline genes of Chiroptera and the pattern of Rhinolophus affinis bat IGH CDR3 repertoire
- PMID: 38465979
- PMCID: PMC10986613
- DOI: 10.1128/spectrum.03762-23
Landscape of IGH germline genes of Chiroptera and the pattern of Rhinolophus affinis bat IGH CDR3 repertoire
Abstract
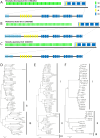
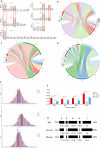
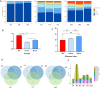
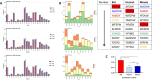
The emergence and re-emergence of abundant viruses from bats that impact human and animal health have resulted in a resurgence of interest in bat immunology. Characterizing the immune receptor repertoire is critical to understanding how bats coexist with viruses in the absence of disease and developing new therapeutics to target viruses in humans and susceptible livestock. In this study, IGH germline genes of Chiroptera including Rhinolophus ferrumequinum, Phyllostomus discolor, and Pipistrellus pipistrellus were annotated, and we profiled the characteristics of Rhinolophus affinis (RA) IGH CDR3 repertoire. The germline genes of Chiroptera are quite different from those of human, mouse, cow, and dog in evolution, but the three bat species have high homology. The CDR3 repertoire of RA is unique in many aspects including CDR3 subclass, V/J genes access and pairing, CDR3 clones, and somatic high-frequency mutation compared with that of human and mouse, which is an important point in understanding the asymptomatic nature of viral infection in bats. This study unveiled a detailed map of bat IGH germline genes on chromosome level and provided the first immune receptor repertoire of bat, which will stimulate new avenues of research that are directly relevant to human health and disease.IMPORTANCEThe intricate relationship between bats and viruses has been a subject of study since the mid-20th century, with more than 100 viruses identified, including those affecting humans. While preliminary investigations have outlined the innate immune responses of bats, the role of adaptive immunity remains unclear. This study presents a pioneering contribution to bat immunology by unveiling, for the first time, a detailed map of bat IGH germline genes at the chromosome level. This breakthrough not only provides a foundation for B cell receptor research in bats but also contributes to primer design and sequencing of the CDR3 repertoire. Additionally, we offer the first comprehensive immune receptor repertoire of bats, serving as a crucial library for future comparative analyses. In summary, this research significantly advances the understanding of bats' immune responses, providing essential resources for further investigations into viral tolerance and potential zoonotic threats.
Keywords: IGH; bat; germline gene; immune repertoire.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
New insights into the germline genes and CDR3 repertoire of the TCRβ chain in Chiroptera.Front Immunol. 2023 Mar 27;14:1147859. doi: 10.3389/fimmu.2023.1147859. eCollection 2023. Front Immunol. 2023. PMID: 37051236 Free PMC article.
-
Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence.Elife. 2020 Feb 3;9:e48401. doi: 10.7554/eLife.48401. Elife. 2020. PMID: 32011232 Free PMC article.
-
Antiviral immune responses of bats: a review.Zoonoses Public Health. 2013 Feb;60(1):104-16. doi: 10.1111/j.1863-2378.2012.01528.x. Epub 2012 Aug 1. Zoonoses Public Health. 2013. PMID: 23302292 Free PMC article. Review.
-
Six reference-quality genomes reveal evolution of bat adaptations.Nature. 2020 Jul;583(7817):578-584. doi: 10.1038/s41586-020-2486-3. Epub 2020 Jul 22. Nature. 2020. PMID: 32699395 Free PMC article.
-
Public health awareness of emerging zoonotic viruses of bats: a European perspective.Vector Borne Zoonotic Dis. 2006 Winter;6(4):315-24. doi: 10.1089/vbz.2006.6.315. Vector Borne Zoonotic Dis. 2006. PMID: 17187565 Review.
Cited by
-
Comparative Analysis of Mammalian Adaptive Immune Loci Revealed Spectacular Divergence and Common Genetic Patterns.Mol Biol Evol. 2025 Jul 1;42(7):msaf152. doi: 10.1093/molbev/msaf152. Mol Biol Evol. 2025. PMID: 40580934 Free PMC article.
-
Annotation and characterization of immunoglobulin loci and CDR3 polymorphism in water buffalo (Bubalus bubalis).Front Immunol. 2025 Jan 20;15:1503788. doi: 10.3389/fimmu.2024.1503788. eCollection 2024. Front Immunol. 2025. PMID: 39902045 Free PMC article.
-
The reverse TRBV30 gene of mammals: a defect or superiority in evolution?BMC Genomics. 2024 Jul 19;25(1):705. doi: 10.1186/s12864-024-10632-4. BMC Genomics. 2024. PMID: 39030501 Free PMC article.
-
Different Species of Bats: Genomics, Transcriptome, and Immune Repertoire.Curr Issues Mol Biol. 2025 Apr 7;47(4):252. doi: 10.3390/cimb47040252. Curr Issues Mol Biol. 2025. PMID: 40699651 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

