Implementation of IFPTML Computational Models in Drug Discovery Against Flaviviridae Family
- PMID: 38466369
- PMCID: PMC10966645
- DOI: 10.1021/acs.jcim.3c01796
Implementation of IFPTML Computational Models in Drug Discovery Against Flaviviridae Family
Abstract
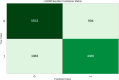
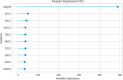
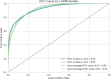
The Flaviviridae family consists of single-stranded positive-sense RNA viruses, which contains the genera Flavivirus, Hepacivirus, Pegivirus, and Pestivirus. Currently, there is an outbreak of viral diseases caused by this family affecting millions of people worldwide, leading to significant morbidity and mortality rates. Advances in computational chemistry have greatly facilitated the discovery of novel drugs and treatments for diseases associated with this family. Chemoinformatic techniques, such as the perturbation theory machine learning method, have played a crucial role in developing new approaches based on ML models that can effectively aid drug discovery. The IFPTML models have shown its capability to handle, classify, and process large data sets with high specificity. The results obtained from different models indicates that this methodology is proficient in processing the data, resulting in a reduction of the false positive rate by 4.25%, along with an accuracy of 83% and reliability of 92%. These values suggest that the model can serve as a computational tool in assisting drug discovery efforts and the development of new treatments against Flaviviridae family diseases.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
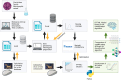
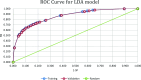
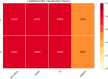
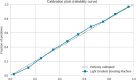
Similar articles
-
Characterization of Recombinant Flaviviridae Viruses Possessing a Small Reporter Tag.J Virol. 2018 Jan 2;92(2):e01582-17. doi: 10.1128/JVI.01582-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29093094 Free PMC article.
-
Current approaches in antiviral drug discovery against the Flaviviridae family.Curr Pharm Des. 2014;20(21):3428-44. doi: 10.2174/13816128113199990635. Curr Pharm Des. 2014. PMID: 24001228 Review.
-
A New Subclass of Exoribonuclease-Resistant RNA Found in Multiple Genera of Flaviviridae.mBio. 2020 Sep 29;11(5):e02352-20. doi: 10.1128/mBio.02352-20. mBio. 2020. PMID: 32994331 Free PMC article.
-
FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae.Viruses. 2020 Aug 14;12(8):892. doi: 10.3390/v12080892. Viruses. 2020. PMID: 32824044 Free PMC article.
-
Delayed by Design: Role of Suboptimal Signal Peptidase Processing of Viral Structural Protein Precursors in Flaviviridae Virus Assembly.Viruses. 2020 Sep 26;12(10):1090. doi: 10.3390/v12101090. Viruses. 2020. PMID: 32993149 Free PMC article. Review.
Cited by
-
Perturbation-Theory Machine Learning for Multi-Target Drug Discovery in Modern Anticancer Research.Curr Issues Mol Biol. 2025 Apr 25;47(5):301. doi: 10.3390/cimb47050301. Curr Issues Mol Biol. 2025. PMID: 40699700 Free PMC article. Review.
-
In Silico Approach for Early Antimalarial Drug Discovery: De Novo Design of Virtual Multi-Strain Antiplasmodial Inhibitors.Microorganisms. 2025 Jul 9;13(7):1620. doi: 10.3390/microorganisms13071620. Microorganisms. 2025. PMID: 40732129 Free PMC article.
References
-
- Capinera J. L.Encyclopedia of Entomology; Springer Science & Business Media, 2008.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

